HIER vs. PIER: A Strategic Guide to Optimizing Antigen Retrieval for Reproducible IHC
This article provides a comprehensive guide for researchers and drug development professionals on optimizing antigen retrieval, a critical step in immunohistochemistry (IHC).
HIER vs. PIER: A Strategic Guide to Optimizing Antigen Retrieval for Reproducible IHC
Abstract
This article provides a comprehensive guide for researchers and drug development professionals on optimizing antigen retrieval, a critical step in immunohistochemistry (IHC). It covers the foundational science behind epitope masking caused by formalin fixation and explores the two primary retrieval methods: Heat-Induced Epitope Retrieval (HIER) and Proteolytic-Induced Epitope Retrieval (PIER). The scope extends to detailed, actionable protocols, systematic troubleshooting for common artifacts, and a comparative analysis of both methods based on recent scientific evidence to guide method selection for specific antigens and tissue types, ultimately ensuring reliable and reproducible staining outcomes in biomedical research.
The Why Behind the Method: Understanding Epitope Masking and Retrieval Fundamentals
Formalin fixation is a cornerstone of histopathology, providing excellent preservation of tissue architecture that pathologists have relied upon for over a century [1]. However, this same process creates a significant challenge for immunohistochemistry (IHC) by masking epitopes—the specific binding sites recognized by antibodies [2]. The formaldehyde in formalin solution induces protein cross-linking, forming methylene bridges between amino acid residues that alter protein conformation and obscure antigenic sites [2] [1]. This masking effect can lead to false-negative results, weak staining intensity, and compromised data interpretation, presenting a critical obstacle for researchers and drug development professionals requiring accurate protein localization and detection.
The discovery of antigen retrieval techniques in the 1990s revolutionized IHC, enabling researchers to reverse these formalin-induced modifications [3]. Today, the choice between Heat-Induced Epitope Retrieval (HIER) and Proteolytic-Induced Epitope Retrieval (PIER) represents a fundamental methodological decision that directly impacts experimental outcomes. Understanding the chemical basis of formalin fixation and the mechanisms by which retrieval methods counteract its effects is essential for optimizing IHC protocols across diverse tissue types and research applications.
The Chemistry of Formalin Fixation and Epitope Masking
Molecular Mechanisms of Cross-linking
Formalin fixation preserves tissue through a time-dependent chemical process that begins when formaldehyde reacts with reactive amino acid side chains to form hydroxymethyl groups [4] [5]. These highly reactive intermediates subsequently form stable methylene bridges between closely spaced amino groups on adjacent proteins [4]. This cross-linking occurs both within individual protein molecules (intramolecular) and between different proteins (intermolecular), creating a dense network that stabilizes tissue architecture but simultaneously masks antigenic epitopes [4].
The cross-linking process affects protein structure through multiple mechanisms:
- Conformational changes that alter the three-dimensional shape of epitopes [2]
- Steric hindrance from bystander proteins cross-linked in close proximity to the target antigen [4]
- Reversal of protein polarity and changes to electrostatic charges that affect antibody binding [4]
Impact of Fixation Variables on Epitope Masking
The extent of epitope masking depends on several fixation parameters. While prolonged formalin fixation was historically presumed to increase antigen masking, contemporary research demonstrates that most antigens maintain immunoreactivity even after extended fixation periods when appropriate retrieval methods are employed [5]. One comprehensive study evaluating 61 different antigens found that only three (cytokeratin 7, high-molecular-weight cytokeratin, and laminin) showed diminished immunoreactivity after prolonged fixation, while the majority maintained moderate to strong staining for up to 7 weeks [5].
Other critical factors include:
- Fixative pH and concentration - Neutral buffered formalin (4% formaldehyde) is the standard [1]
- Tissue size and permeability - Thin sections fix more uniformly [1]
- Fixation temperature - Room temperature is standard, though some protocols use elevated temperatures [4]
Antigen Retrieval Methodologies: HIER vs. PIER
Heat-Induced Epitope Retrieval (HIER)
HIER utilizes elevated temperatures (typically 95-100°C) to disrupt formalin-induced crosslinks through thermal energy [2] [6]. The mechanism is believed to involve both hydrolytic cleavage of methylene bridges and protein unfolding that restores epitope conformation [6]. Buffer composition and pH critically influence HIER effectiveness by affecting calcium ion chelation and the stability of protein structures during heating [2].
Standard HIER buffers include:
- Sodium citrate buffer (pH 6.0) - Effective for many nuclear and cytoplasmic antigens [6]
- Tris-EDTA buffer (pH 9.0) - Often superior for membrane proteins and challenging targets [2] [6]
- EDTA buffer (pH 8.0) - Useful for certain nuclear antigens [6]
HIER implementation varies by heating method, with each offering distinct advantages:
Table 1: HIER Heating Method Comparison
| Method | Temperature | Time | Advantages | Limitations |
|---|---|---|---|---|
| Pressure Cooker | ~120°C | 3-10 minutes | Rapid, uniform heating | Can damage delicate tissues |
| Microwave | 95-100°C | 15-20 minutes | Widely accessible | Potential hot spots |
| Steamer/Water Bath | 95-100°C | 20-30 minutes | Gentle, consistent | Longer processing time |
| Autoclave | 120°C+ | Short cycles | Standardized conditions | Equipment access required |
Proteolytic-Induced Epitope Retrieval (PIER)
PIER employs proteolytic enzymes to cleave peptide bonds within the cross-linked protein network, physically removing obstructive proteins and exposing masked epitopes [2] [7]. This method operates at milder temperatures (typically 37°C) but requires precise optimization to balance epitope exposure against potential tissue damage [2].
Common PIER enzymes include:
- Proteinase K - Broad-spectrum serine protease effective for many extracellular antigens [8]
- Trypsin - Cleaves carboxyl side of lysine and arginine residues [2]
- Pepsin - Prefers cleavage at phenylalanine, leucine, and tyrosine residues [7]
Critical optimization parameters for PIER include enzyme concentration, digestion time, temperature, and pH, all of which must be empirically determined for each antigen-antibody system [2] [1]. The College of American Pathologists recommends formalin fixation for a minimum of 6 hours and a maximum of 48 hours for optimal results [1].
Comparative Analysis: HIER vs. PIER
Table 2: Direct Comparison of HIER and PIER Methods
| Parameter | HIER | PIER |
|---|---|---|
| Mechanism | Thermal unfolding of crosslinks [9] | Enzymatic cleavage of proteins [9] |
| Typical Conditions | 95-100°C for 10-30 minutes [2] | 37°C for 5-30 minutes [2] |
| Buffer Systems | Citrate (pH 6.0), Tris-EDTA (pH 9.0) [6] | Tris/HCl, PBS (typically neutral pH) [8] [7] |
| Advantages | Higher success rate, gentler on morphology [10] | Effective for select difficult epitopes [2] |
| Disadvantages | Potential tissue detachment [8] | Risk of over-digestion and tissue damage [2] |
| Success Rate | High (>80% of antigens) [10] | Variable (antigen-dependent) [3] |
Experimental Evidence and Comparative Studies
Case Study: CILP-2 Detection in Articular Cartilage
A recent systematic comparison of antigen retrieval methods for detecting cartilage intermediate layer protein 2 (CILP-2) in osteoarthritic cartilage demonstrated the antigen-specific nature of retrieval optimization [8]. Researchers evaluated four different protocols on knee replacement samples, with semi-quantitative assessment revealing that PIER alone provided superior results for this particular extracellular matrix glycoprotein [8].
Notably, the combination of HIER and PIER not only failed to improve staining but actually reduced immunoreactivity and caused frequent section detachment [8]. This finding highlights that more aggressive retrieval does not necessarily yield better outcomes and must be tailored to the specific target. The study attributed PIER's success with CILP-2 to the enzyme's ability to digest the dense cartilage matrix while preserving the less glycosylated (and potentially more heat-labile) CILP-2 epitopes [8].
Broader Patterns in Antigen Retrieval Efficacy
Comprehensive studies across multiple tissue types have revealed that most antigens benefit from HIER, with buffer pH representing a critical variable [5] [10]. However, certain antigen classes show distinct preferences:
- Nuclear antigens (e.g., Ki-67, estrogen receptor) typically respond best to high-pHIER [4] [5]
- Cytoplasmic intermediate filaments often require either HIER or enzymatic methods depending on the specific protein [5]
- Extracellular matrix components (particularly in dense tissues) may benefit from PIER or combined approaches [8]
- Membrane proteins show variable responses, with many detecting best with high-pH buffers [5]
Research has demonstrated that for the majority of antigens (approximately 95% in one comprehensive study), immunoreactivity remains detectable even after prolonged formalin fixation (up to 7 weeks) when appropriate retrieval methods are applied [5].
Detailed Experimental Protocols
Standardized HIER Protocol Using Pressure Cooker
This protocol adapts methods from multiple sources to provide a robust starting point for HIER optimization [6] [10]:
Reagents and Equipment:
- Domestic stainless steel pressure cooker
- Hot plate
- Slide rack (metal or plastic)
- Antigen retrieval buffer (citrate pH 6.0, Tris-EDTA pH 9.0, or EDTA pH 8.0)
Procedure:
- Prepare 400-500 mL of selected antigen retrieval buffer in pressure cooker
- Place pressure cooker on hot plate at full power without securing lid
- While buffer heats, deparaffinize and rehydrate tissue sections using standard xylene/ethanol series
- Once buffer reaches boiling, transfer slides from hydration bath to pressure cooker using forceps
- Secure pressure cooker lid according to manufacturer instructions
- Once full pressure is achieved, time exactly 3 minutes
- After 3 minutes, turn off hot plate and transfer pressure cooker to sink
- Activate pressure release valve and run cold water over cooker for 10 minutes
- Carefully open lid and run cold tap water into cooker for additional 10 minutes
- Proceed with standard immunohistochemical staining protocol
Optimization Notes: For delicate tissues (cartilage, skin), reduce pressure time to 1-2 minutes or use water bath method (60°C overnight) to prevent section loss [6].
Standardized PIER Protocol Using Proteinase K
This protocol is adapted from cartilage matrix protein research with general applicability [8]:
Reagents:
- Proteinase K (30 µg/mL in 50 mM Tris/HCl, 5 mM CaCl₂, pH 6.0)
- Hyaluronidase (0.4% in HEPES-buffered medium)
- Humidified incubation chamber
Procedure:
- Deparaffinize and rehydrate tissue sections through xylene and graded ethanol series
- Wash slides in distilled water for 5 minutes
- Apply Proteinase K solution to completely cover tissue sections
- Incubate in humidified chamber at 37°C for 90 minutes
- Rinse slides gently in PBS (pH 7.4) for 3 × 5 minutes
- Apply hyaluronidase solution to sections
- Incubate in humidified chamber at 37°C for 3 hours
- Rinse thoroughly in PBS for 3 × 5 minutes
- Proceed with peroxidase blocking and standard IHC protocol
Optimization Notes: Proteinase K concentration (10-50 µg/mL) and incubation time (30-120 minutes) should be titrated based on fixation duration and tissue type [8] [1].
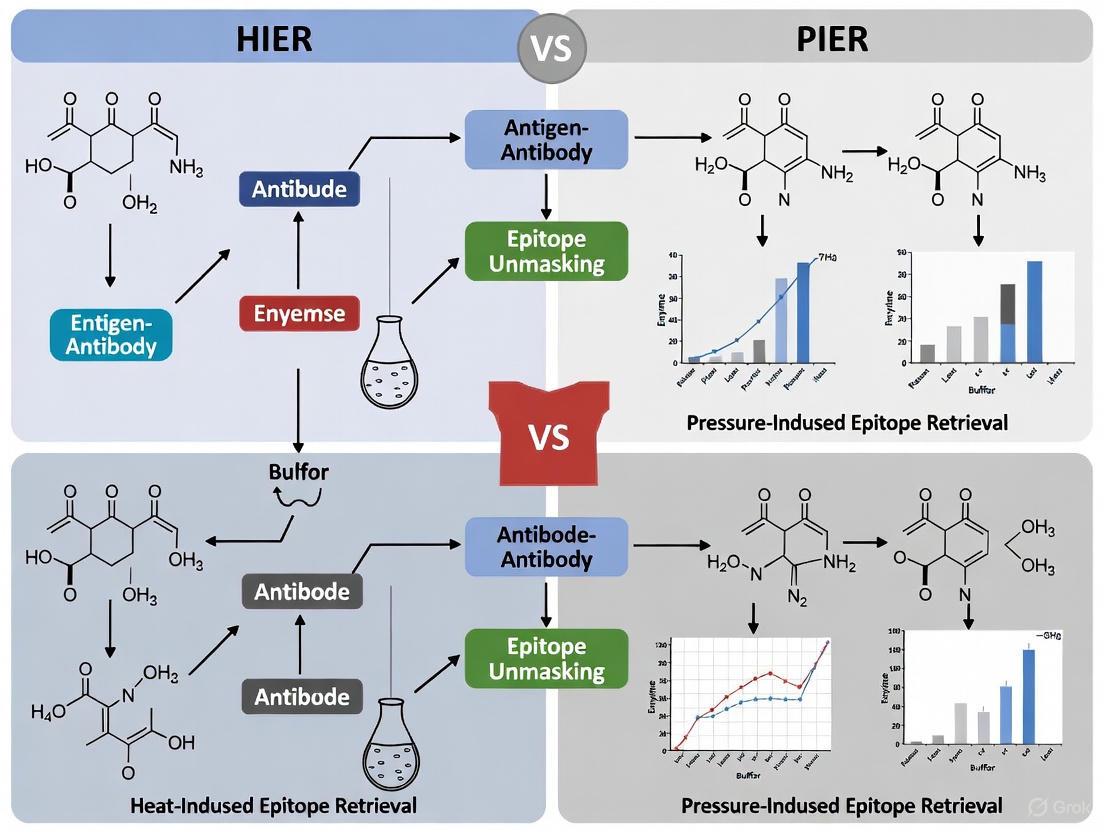
Visualization of Experimental Workflows and Method Selection
Diagram 1: Antigen Retrieval Method Selection Algorithm - A systematic workflow for selecting and optimizing antigen retrieval methods based on experimental needs and preliminary results.
Diagram 2: HIER vs. PIER - Mechanisms and Outcomes - Comparative visualization of the fundamental processes and resulting effects of both antigen retrieval methodologies.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Essential Reagents for Antigen Retrieval Optimization
| Reagent/Equipment | Function/Purpose | Examples/Notes |
|---|---|---|
| Sodium Citrate Buffer | Low-pH (6.0) retrieval solution | Effective for many nuclear antigens [6] |
| Tris-EDTA Buffer | High-pH (9.0) retrieval solution | Preferred for many membrane proteins [6] |
| Proteinase K | Serine protease for PIER | 10-50 µg/mL in Tris/HCl with CaCl₂ [8] |
| Trypsin | Protease for alternative PIER | 0.1% solution in PBS or Tris buffer [2] |
| Pressure Cooker | HIER heating device | Provides uniform heating at ~120°C [6] |
| Scientific Microwave | Alternative HIER device | Maintains constant 98°C for 20 minutes [6] |
| Humidified Chamber | PIER incubation | Maintains enzyme activity during 37°C incubation [8] |
| Positive Control Tissues | Validation of protocol effectiveness | Tissues with known antigen expression [2] |
The formalin fixation problem represents a fundamental challenge in immunohistochemistry, but comprehensive understanding of cross-linking mechanisms and antigen retrieval techniques enables researchers to overcome these limitations effectively. The strategic selection between HIER and PIER, coupled with systematic optimization of critical parameters including buffer pH, temperature, and duration, enables successful epitope unmasking for the vast majority of targets.
Future directions in antigen retrieval research include:
- Development of target-specific retrieval cocktails that combine thermal and enzymatic approaches
- Automated standardization of retrieval conditions across laboratory platforms
- Computational prediction of optimal retrieval methods based on antigen physicochemical properties
- Novel chemical approaches beyond traditional HIER and PIER for particularly challenging epitopes
As IHC continues to evolve as a critical tool in both basic research and clinical diagnostics, the principles outlined in this application note provide a foundation for robust, reproducible antigen detection across diverse experimental systems.
Formalin fixation and paraffin embedding (FFPE) is the gold standard for preserving tissue morphology in diagnostic pathology and research. However, a significant drawback of this process is the masking of antigenic epitopes, which can impair antibody binding and compromise the sensitivity and specificity of immunohistochemistry (IHC) [11] [2]. The core principle of antigen retrieval is to reverse these formalin-induced chemical modifications, thereby restoring immunoreactivity and enabling accurate protein detection in FFPE tissues [3].
Formalin fixation primarily works by creating methylene bridges between protein molecules [11]. Specifically, formaldehyde reacts with amino acid side chains to form hydroxymethyl groups, which then cross-link with other tissue proteins over hours to days [12]. These cross-links can sterically block antibody access to epitopes and alter protein conformation, effectively "masking" antigens from detection [12] [2]. The discovery of antigen retrieval in 1991 represented a milestone in IHC, making it possible to reliably detect antigens in FFPE tissues that were previously inaccessible [3].
Fundamental Mechanisms: How Antigen Retrieval Reverses Formalin's Effects
Molecular Principles of Epitope Masking and Unmasking
The prevailing understanding of antigen retrieval mechanisms involves the reversal of protein cross-links formed during formalin fixation [3]. Research using peptide epitope mapping has demonstrated that most clinically useful antibodies for FFPE tissues recognize linear epitopes—contiguous stretches of amino acids in the native protein—rather than conformational epitopes that depend on three-dimensional protein folding [12]. When formalin fixation occurs in the presence of irrelevant proteins, these can become cross-linked to the peptide epitopes, creating steric hindrance that prevents antibody binding [12]. Antigen retrieval works by dissociating these irrelevant proteins and restoring antibody access [12].
The mechanism of action differs between heat-induced and protease-induced methods:
HIER utilizes high temperatures (95-100°C) to disrupt the weaker non-covalent bonds and potentially reverse some formalin-induced cross-links through thermal unfolding [8] [2]. The chemical composition and pH of the retrieval buffer significantly influence this process, with some buffers possibly acting through calcium ion chelation [13] [2].
PIER employs proteolytic enzymes (e.g., proteinase K, trypsin) to selectively digest proteins surrounding the epitopes, effectively cleaving the cross-links that mask antigenic sites [8] [13]. This method is generally considered gentler on tissues but carries the risk of destroying the antigen of interest if not carefully optimized [13] [3].
Visualizing the Process of Antigen Retrieval
The following diagram illustrates the core principle of how formalin fixation masks epitopes and how antigen retrieval reverses this effect:
Visualizing Antigen Retrieval Principle
This fundamental process of unmasking epitopes enables researchers to overcome the primary challenge of working with FFPE tissues and forms the basis for all antigen retrieval methodologies discussed in this application note.
Methodological Comparison: HIER versus PIER
Technical Specifications and Applications
The two primary antigen retrieval methods—Heat-Induced Epitope Retrieval (HIER) and Proteolytic-Induced Epitope Retrieval (PIER)—differ significantly in their mechanisms, applications, and optimization requirements. The table below provides a comparative analysis of these methods:
Table 1: Comprehensive Comparison of HIER and PIER Methods
| Parameter | Heat-Induced Epitope Retrieval (HIER) | Proteolytic-Induced Epitope Retrieval (PIER) |
|---|---|---|
| Mechanism of Action | Thermal disruption of protein cross-links through high-temperature heating [13] [2] | Enzymatic cleavage of cross-linking proteins using proteases [8] [13] |
| Typical Conditions | 95-100°C for 10-30 minutes in specific buffer solutions [6] [13] | 37°C for 10-30 minutes with enzyme-specific buffers [13] [2] |
| Common Reagents | Citrate buffer (pH 6.0), Tris-EDTA (pH 9.0), EDTA (pH 8.0) [6] [13] | Proteinase K, trypsin, pepsin, pronase [8] [13] |
| Equipment | Pressure cooker, microwave, vegetable steamer, water bath [6] [13] | Incubator, humidified chamber [13] |
| Primary Advantages | Suitable for a broader range of antigens [13]; Better preservation of tissue morphology [13]; Less non-specific staining [13] | Preferred for difficult-to-recover epitopes [13]; Less damaging to delicate tissues [13]; Effective for certain antigens in dense matrices [8] |
| Key Limitations | Potential for tissue damage or antigenicity loss with overheating [8] [13]; Section detachment issues [8] [14] | Narrow optimal concentration range [13] [2]; Potential destruction of tissue morphology and antigens [13] [3]; Lower success rate for restoring immunoreactivity [3] |
| Ideal Applications | Nuclear antigens [13]; High-molecular-weight proteins [13]; General screening purposes [2] | Fragile tissues [13]; Cartilage matrix proteins [8]; Epitopes resistant to heat retrieval [8] |
Experimental Evidence and Performance Data
Recent comparative studies have provided quantitative data on the performance of HIER versus PIER across different tissue types and experimental conditions:
Table 2: Experimental Performance Comparison of HIER and PIER Across Tissue Types
| Tissue Type / Antigen | Best Performing Method | Performance Metrics | Study Reference |
|---|---|---|---|
| Osteoarthritic cartilage (CILP-2) | PIER (Proteinase K + hyaluronidase) | Most abundant staining; HIER resulted in frequent section detachment [8] | Methods Protoc. 2024 |
| Mouse decalcified joint tissues (p65, IL-1β) | Trypsin retrieval | Better tissue morphology preservation; IWB method showed higher positive signal percentage but more detachment [14] | J Immunol Methods 2023 |
| Murine female reproductive tract (ECP, HSV-2) | HIER (80°C in citrate buffer) | Increased antibody binding; best tissue morphology; most efficient for automated analysis [15] | Eur J Histochem 2024 |
| General IHC applications | HIER (majority of cases) | Effective for >90% of antigens; preferred for nuclear antigens with high-pH buffers [13] [2] | Commercial Protocols |
The tissue-specific performance variations highlighted in these studies underscore the importance of empirical optimization rather than relying on universal protocols.
Experimental Protocols and Optimization Strategies
Detailed HIER Protocol Using Microwave Method
The microwave HIER method provides a balance of efficiency and consistency for most laboratory settings:
Materials Required: Microwave (domestic or scientific), microwaveable vessel with slide rack, antigen retrieval buffer (e.g., Tris-EDTA pH 9.0, sodium citrate pH 6.0) [6]
Buffer Preparation:
- Sodium citrate buffer (10 mM, pH 6.0): 2.94 g tri-sodium citrate (dihydrate) in 1L distilled water, adjust to pH 6.0 with HCl, add 0.5 mL Tween 20 [6]
- Tris-EDTA buffer (10 mM Tris, 1 mM EDTA, pH 9.0): 1.21 g Tris base, 0.37 g EDTA in 1L distilled water, adjust to pH 9.0, add 0.5 mL Tween 20 [6]
- EDTA buffer (1 mM, pH 8.0): 0.37 g EDTA in 1L distilled water, adjust to pH 8.0 with NaOH [6]
Step-by-Step Procedure:
- Deparaffinize and rehydrate tissue sections using standard protocols [6]
- Place slides in microwaveable vessel containing sufficient antigen retrieval buffer to cover slides by at least a few centimeters [6]
- Microwave at 95°C for 8 minutes, then cool slides for 5 minutes [13]
- Microwave again at 95°C for 4 minutes, then cool to room temperature [13]
- Continue with standard IHC staining protocol [6]
Critical Optimization Parameters:
Detailed PIER Protocol Using Proteinase K
For antigens resistant to heat retrieval or in specialized tissues, the enzymatic approach may be superior:
Materials Required: Proteinase K (or alternative: trypsin, pepsin), 37°C incubator, humidified chamber, Tris/HCl buffer [8] [15]
Enzyme Solution Preparation:
Step-by-Step Procedure:
- Deparaffinize and rehydrate tissue sections through xylene and graded ethanol series [8]
- Pipette Proteinase K working solution onto tissue sections [15]
- Place slides in humidity chamber and incubate for 15-90 minutes at 37°C [8] [15]
- Transfer slides to tap water and rinse for 3-5 minutes [13]
- Continue with standard IHC staining protocol [8]
Critical Optimization Parameters:
- Enzyme Concentration: Titrate from 10-50 µg/mL for Proteinase K to balance epitope exposure versus tissue preservation [8] [15]
- Incubation Time: Test between 10-30 minutes for most enzymes; extend for difficult epitopes [8] [13]
- Tissue Considerations: Cartilage and other dense matrices may require longer incubation or combined enzymatic approaches [8]
Systematic Optimization Strategy
A structured approach to antigen retrieval optimization significantly enhances IHC outcomes:
- Initial Screening Matrix: Test a combination of retrieval methods and buffer pH values using a systematic approach [13]:
Table 3: Antigen Retrieval Optimization Matrix Template
| Time | Citrate Buffer (pH 6.0) | Tris-EDTA (pH 8.0) | Tris-EDTA (pH 9.0) |
|---|---|---|---|
| 8 minutes | Slide #1 | Slide #2 | Slide #3 |
| 15 minutes | Slide #4 | Slide #5 | Slide #6 |
| 20 minutes | Slide #7 | Slide #8 | Slide #9 |
- Troubleshooting Common Issues:
- Weak or No Staining: Increase heating time, switch to higher pH buffer, or try enzymatic retrieval [2]
- Tissue Detachment: Use poly-L-lysine coated slides, reduce heating time, or switch to enzymatic method [8] [14]
- High Background: Reduce heating time, decrease enzyme concentration, or optimize antibody dilution [2]
Research Reagent Solutions and Equipment
Successful antigen retrieval implementation requires specific reagents and equipment. The following table details essential solutions and their applications:
Table 4: Essential Research Reagents for Antigen Retrieval Optimization
| Reagent / Equipment | Specification / Composition | Primary Function | Application Notes |
|---|---|---|---|
| Citrate Buffer | 10 mM sodium citrate, 0.05% Tween 20, pH 6.0 [6] | Low-pHIER buffer; effective for many cytoplasmic antigens [13] | Traditional standard; less effective for nuclear antigens [13] |
| Tris-EDTA Buffer | 10 mM Tris base, 1 mM EDTA, 0.05% Tween 20, pH 9.0 [6] | High-pH HIER buffer; particularly effective for nuclear antigens [13] | Increasingly preferred over citrate for broader antigen range [13] |
| EDTA Buffer | 1 mM EDTA, pH 8.0 [6] | Chelating buffer for HIER; calcium ion extraction [13] [2] | Suitable for broad antigen range with minimal morphological damage [13] |
| Proteinase K | 30 µg/mL in 50 mM Tris/HCl with 5 mM CaCl₂ (pH 6.0) [8] | Proteolytic enzyme for PIER; cleaves peptide bonds [8] [15] | Optimal for dense matrices like cartilage; requires concentration optimization [8] |
| Trypsin | 0.1% trypsin in appropriate buffer [13] | Proteolytic enzyme for PIER; specific cleavage at lysine/arginine [13] | Standard enzymatic retrieval; incubation typically 10-30 minutes at 37°C [13] |
| Pressure Cooker | Domestic stainless steel pressure cooker [6] | HIER equipment providing consistent high temperature under pressure [6] | Rapid heating (2-3 minutes at full pressure); minimizes section detachment [6] |
| Scientific Microwave | Temperature-controlled microwave system [6] | HIER equipment with precise temperature regulation [6] | Preferable to domestic microwaves due to uniform heating [6] |
Antigen retrieval remains a cornerstone technique for modern immunohistochemistry, enabling researchers to overcome the fundamental challenge of epitope masking in FFPE tissues. The core principle—reversing formalin-induced crosslinks through either physical (HIER) or chemical (PIER) means—has proven remarkably durable since its discovery three decades ago.
The comparative data presented in this application note demonstrates that method selection must be antigen-specific and tissue-dependent. While HIER generally offers broader applicability, PIER shows superior performance for specific applications, particularly in challenging tissues like cartilage [8] or when dealing with heat-labile epitopes. The ongoing refinement of antigen retrieval protocols, including the development of specialized buffers and equipment, continues to enhance the sensitivity and reproducibility of IHC across diverse research and diagnostic applications.
Future directions in antigen retrieval methodology may include the integration of computational approaches for protocol optimization [16] and the development of standardized controls for quality assurance [12]. As antibody-based technologies advance, including the application of deep learning for antibody optimization [16], the fundamental principle of antigen retrieval—reversing formalin's effects to reveal hidden epitopes—will remain essential to unlocking the full potential of immunohistochemistry in research and diagnostic applications.
The evolution of antigen retrieval techniques represents a pivotal chapter in the history of immunohistochemistry (IHC), fundamentally transforming how researchers detect proteins in formalin-fixed tissues. For decades, the proteolytic-induced epitope retrieval (PIER) method served as the sole approach for unmasking antigens obscured by formaldehyde fixation [17]. This landscape underwent a dramatic shift in the 1990s with the introduction of heat-induced epitope retrieval (HIER), a revolutionary development that expanded the capabilities of IHC and enhanced detection sensitivity for numerous antigens [18]. The transition from PIER to HIER did not render enzymatic methods obsolete but rather established a diversified toolkit that researchers must strategically navigate based on their specific experimental needs [19] [20]. This application note examines the historical context of these methodologies and provides contemporary protocols framed within a thesis dedicated to optimizing antigen retrieval for advanced research applications.
The PIER Era: Foundations of Antigen Retrieval
The mid-1970s marked the emergence of proteolytic-induced epitope retrieval (PIER) as the first systematic approach to reversing formaldehyde-induced crosslinks in tissue specimens [17]. This technique relies on enzymatic digestion to degrade proteins that mask epitopes, thereby restoring antibody accessibility. The fundamental principle involves using proteases such as trypsin, proteinase K, pronase, ficin, and pepsin to break the methylene bridges formed during formalin fixation [17] [6]. The effectiveness of PIER depends on multiple factors including enzyme concentration and type, incubation parameters (time, temperature, and pH), and fixation duration [17].
Table 1: Common Enzymes Used in PIER Protocols
| Enzyme | Typical Concentration | Incubation Conditions | Primary Applications |
|---|---|---|---|
| Proteinase K | 20 μg/mL [21] | 10-20 min at 37°C [21] | Cartilage proteins, cross-linked antigens [17] |
| Trypsin | 0.05% [21] | 10-20 min at 37°C [21] | General tissue antigens |
| Pepsin | 0.1-0.5% [6] | 10-30 min at 37°C [6] | Extracellular matrix targets |
Despite its pioneering status, PIER presents significant limitations, including potential damage to tissue morphology, alteration of antigen integrity, and technical demanding protocols [17] [22]. These constraints prompted the investigation of alternative methods, setting the stage for a paradigm shift in antigen retrieval approaches.
The HIER Revolution: A Technical Transformation
The 1990s witnessed a revolutionary advancement with the introduction of heat-induced epitope retrieval (HIER), which dramatically enhanced the sensitivity of IHC for formalin-fixed tissues [18]. This technique utilizes high temperatures in combination with specific buffered solutions to reverse formaldehyde-mediated chemical modifications of antigens [18]. The exact mechanism remains partially elucidated, but leading theories suggest that thermal energy breaks protein crosslinks [18] and calcium chelation in specific buffers removes calcium ions from crosslink sites [18].
The implementation of HIER involves multiple heating platforms, each with distinct advantages and limitations:
Table 2: Comparison of HIER Heating Platforms
| Heating Source | Temperature Range | Advantages | Disadvantages |
|---|---|---|---|
| Pressure Cooker | 110-120°C [18] | Short time, high sensitivity [18] | Potential tissue artifacts [18] |
| Vegetable Steamer | 95-100°C [6] [18] | Even heat, good morphology [18] | Longer heating time [18] |
| Water Bath | 92-95°C [23] | Even heat distribution [18] | Longer heating time, expensive [18] |
| Microwave | 94-100°C [6] [18] | Rapid heating, inexpensive [18] | Uneven retrieval, tissue detachment [18] |
The development of HIER also introduced the critical importance of retrieval buffer chemistry. Current evidence indicates that buffer pH significantly influences retrieval effectiveness, with optimal recovery for most epitopes occurring in alkaline buffers (pH 8-10) [18]. EDTA-based buffers are particularly effective for over-fixed specimens and challenging antigens, though they may compromise morphology [18].
Methodological Comparison: An Osteoarthritic Cartilage Case Study
A recent investigation comparing antigen retrieval methods for detecting cartilage intermediate layer protein 2 (CILP-2) in osteoarthritic cartilage provides valuable insights for methodological selection [17] [24]. This study evaluated four protocols: HIER alone, PIER alone (using proteinase K and hyaluronidase), combined HIER/PIER, and no retrieval (control) [17].
The findings demonstrated superior performance of PIER alone for CILP-2 detection, underscoring the persistent relevance of enzymatic methods for specific applications [17] [24]. Contrary to theoretical expectations, combining HIER with PIER not only failed to improve staining but actually diminished the positive effects of enzymatic treatment alone while increasing technical complications such as tissue detachment from slides [17].
Table 3: Semi-Quantitative Staining Assessment for CILP-2 Detection
| Retrieval Method | Staining Extent | Tissue Preservation | Technical Reliability |
|---|---|---|---|
| No Retrieval (Control) | Minimal [17] | Excellent [17] | High [17] |
| HIER Only | Moderate [17] | Good [17] | Moderate [17] |
| PIER Only | Maximal [17] | Moderate [17] | High [17] |
| HIER/PIER Combined | Reduced vs. PIER [17] | Poor (frequent detachment) [17] | Low [17] |
This case study highlights the protein-specific nature of optimal retrieval conditions and emphasizes that advanced glycoproteins in dense matrices like cartilage may respond differently to various retrieval strategies [17].
Detailed Experimental Protocols
Proteolytic-Induced Epitope Retrieval (PIER) Protocol
Materials Required:
- Proteinase K (20 μg/mL working solution) [21] or Trypsin (0.05% working solution) [21]
- TE Buffer, pH 8.0 (for Proteinase K) or Calcium chloride solution (for Trypsin) [21]
- Humidified incubation chamber
- APES-coated slides [22]
Procedure:
- Deparaffinize and rehydrate tissue sections through xylene and graded ethanol series [17] [22]
- Prepare Proteinase K working solution (20 μg/mL) by diluting stock solution in TE Buffer [21]
- Cover tissue sections completely with enzyme solution
- Incubate for 10-20 minutes at 37°C in a humidified chamber [21]
- Allow sections to cool at room temperature for 10 minutes [21]
- Proceed with standard IHC staining protocol [17]
Optimization Notes: Incubation time requires empirical adjustment based on tissue type and fixation duration [21]. Excessive digestion can compromise morphology, while insufficient treatment may yield suboptimal antigen retrieval [17].
Heat-Induced Epitope Retrieval (HIER) Protocol
Materials Required:
- Antigen retrieval buffer (citrate pH 6.0, Tris-EDTA pH 9.0, or EDTA pH 8.0) [6]
- Heating apparatus (pressure cooker, steamer, water bath, or scientific microwave)
- Slide rack and appropriate vessel
- APES-coated slides to prevent detachment [22]
Procedure (Pressure Cooker Method):
- Deparaffinize and rehydrate tissue sections [6]
- Add antigen retrieval buffer to pressure cooker [6]
- Heat buffer to boiling on hot plate [6]
- Transfer slides to boiling buffer [6]
- Secure lid and maintain at full pressure for 3 minutes [6]
- Depressurize rapidly and cool by running cold water over cooker [6]
- Open lid carefully and run cold water into cooker for 10 minutes [6]
- Continue with standard IHC protocol [6]
Alternative Methods:
- Water Bath: Incubate slides in preheated retrieval solution at 92-95°C for 2-10 minutes [23]
- Microwave: Boil slides in retrieval solution for 15-20 minutes, monitoring for evaporation [6] [22]
- Vegetable Steamer: Maintain slides at 95-100°C for 20 minutes [6]
The Scientist's Toolkit: Research Reagent Solutions
Table 4: Essential Reagents for Antigen Retrieval Optimization
| Reagent/Category | Specific Examples | Function & Application Notes |
|---|---|---|
| HIER Buffers | Citrate (pH 6.0), Tris-EDTA (pH 9.0), EDTA (pH 8.0) [6] | Breaks protein crosslinks; pH critical for efficacy [18] |
| PIER Enzymes | Proteinase K, Trypsin, Pepsin [6] [21] | Digests masking proteins; concentration requires optimization [17] |
| Heating Equipment | Pressure cooker, steamer, water bath, scientific microwave [18] | Provides thermal energy for HIER; choice affects time/temperature balance [18] |
| Specialized Slides | APES-coated slides [22] | Prevents tissue detachment during rigorous retrieval procedures [17] [22] |
| Retrieval Kits | Commercial universal retrieval kits [6] [23] | Standardized solutions for reproducible results across multiple antigens |
Strategic Decision Framework
Selecting between PIER and HIER requires systematic consideration of multiple factors. The following workflow provides a logical decision pathway for researchers designing antigen retrieval strategies:
This decision framework emphasizes beginning with literature review and manufacturer recommendations before proceeding to empirical testing [19]. For antigens without established protocols, HIER with neutral buffers typically serves as the appropriate starting point [19]. Should initial results prove unsatisfactory, researchers should systematically explore alternative pH conditions, heating durations, and temperatures before considering PIER approaches [23] [19].
The historical transition from PIER to HIER represents more than mere technical progression—it embodies the evolving sophistication of antigen retrieval methodology. Rather than establishing HIER as a universal replacement for PIER, three decades of application have revealed a nuanced reality where both methods maintain distinct roles within the researcher's arsenal. The contemporary challenge lies not in identifying a singular superior technique but in recognizing the protein-specific, tissue-dependent, and fixation-influenced factors that dictate optimal retrieval conditions. As demonstrated by the CILP-2 case study, certain proteins in challenging matrices like cartilage may respond optimally to traditional enzymatic methods despite the widespread preference for heat-induced approaches [17] [24]. This historical context underscores the enduring importance of methodological optimization and empirical validation in advancing immunohistochemical research.
When is Retrieval Necessary? (FFPE vs. Frozen Tissue Considerations)
Immunohistochemistry (IHC) is an indispensable technique that allows researchers and pathologists to visualize protein distribution, subcellular localization, and abundance within tissue architecture [11]. The foundation of successful IHC lies in the effective binding of antibodies to their specific target epitopes. However, the very process of tissue preservation, essential for maintaining morphological integrity, can significantly impede this antibody-antigen interaction [2]. The necessity for antigen retrieval is therefore intrinsically linked to the method of tissue preservation employed.
Formalin-fixed paraffin-embedded (FFPE) and frozen tissue sections represent the two primary archival methods in biomedical research and clinical diagnostics [25] [26]. A fundamental understanding of how these preparation techniques differently affect epitope accessibility is crucial for designing robust and reproducible IHC experiments. This application note delineates the specific circumstances under which antigen retrieval is necessary, providing a structured framework for researchers to optimize their immunohistochemical protocols, with a particular focus on the comparison between Heat-Induced Epitope Retrieval (HIER) and Proteolytic-Induced Epitope Retrieval (PIER) methodologies.
FFPE vs. Frozen Tissues: Fundamental Differences Driving Retrieval Requirements
The choice between FFPE and frozen tissue preservation creates a fundamental divergence in the workflow and requirements for successful IHC, primarily due to the impact of fixation on protein structure.
Formalin-Fixed Paraffin-Embedded (FFPE) Tissues: Formalin fixation works by creating methylene bridges (-CH2-) that cross-link proteins and nucleic acids within the tissue. This process excellently preserves tissue morphology but alters the three-dimensional conformation of proteins, often masking the epitopes recognized by antibodies [11] [2]. This masking is the primary reason antigen retrieval is typically necessary for FFPE tissues. The cross-linking must be reversed to expose the hidden epitopes for antibody binding.
Frozen Tissues: Frozen tissues are typically "flash-frozen" in liquid nitrogen and stored at -80°C. This process preserves proteins in their native, non-denatured state. While some brief fixation (e.g., with ice-cold acetone) may be performed post-sectioning, it does not create the extensive cross-linking seen with formalin [2] [27]. Consequently, antigen retrieval is generally not required for frozen sections, as the epitopes remain naturally accessible.
Table 1: Core Characteristics of FFPE and Frozen Tissues Influencing Antigen Retrieval
| Characteristic | FFPE Tissues | Frozen Tissues |
|---|---|---|
| Primary Fixative | Formalin (Cross-linking) | Acetone, Methanol, or Ethanol (Precipitative) [11] [27] |
| Effect on Proteins | Denatured, cross-linked | Native structure largely preserved [26] |
| Epitope Status | Often masked | Typically accessible [2] |
| Antigen Retrieval | Usually Required | Usually Not Required [2] [27] |
| Key Advantage | Superior morphology; vast archived biobanks [25] [26] | Optimal for labile epitopes and nucleic acid analysis [25] [26] |
Antigen Retrieval Methodologies: HIER vs. PIER
To overcome the challenge of epitope masking in FFPE tissues, two principal antigen retrieval methods have been developed: Heat-Induced Epitope Retrieval (HIER) and Proteolytic-Induced Epitope Retrieval (PIER).
Heat-Induced Epitope Retrieval (HIER)
Principle: HIER uses high-temperature heating (typically 95-100°C) in a specific buffer solution to break the methylene cross-links formed during formalin fixation. This process thermally disrupts the crosslinks, allowing epitopes to revert to their original conformation [2] [28].
Key Optimization Parameters:
- Buffer pH: The pH of the retrieval buffer is critical. Common buffers include Sodium Citrate (pH 6.0, acidic), PBS (pH 7.2-7.6, neutral), and Tris-EDTA (pH 8.0-9.0, basic) [6] [29]. The optimal pH is epitope-dependent and must be determined empirically.
- Heating Method & Time: Methods include pressure cookers (e.g., 3 min at full pressure), microwaves (e.g., 20 min at 98°C), steamers, and water baths [6]. Higher temperatures generally allow for shorter incubation times.
Proteolytic-Induced Epitope Retrieval (PIER)
Principle: PIER employs proteolytic enzymes such as Proteinase K, trypsin, or pepsin to cleave the protein cross-links and unmask the epitopes [2] [8].
Key Optimization Parameters:
- Enzyme Type and Concentration: Different enzymes have different cleavage specificities.
- Incubation Time and Temperature: PIER is typically performed at 37°C for 10-20 minutes [2]. Over-digestion can destroy the epitope and damage tissue morphology.
Table 2: Comparison of HIER and PIER Methodologies
| Parameter | Heat-Induced Epitope Retrieval (HIER) | Proteolytic-Induced Epitope Retrieval (PIER) |
|---|---|---|
| Principle | Physical reversal of cross-links via heat [2] [28] | Chemical cleavage of cross-links via enzymatic digestion [2] [8] |
| Typical Conditions | 95-100°C for 10-20 min in buffer [6] [29] | 37°C for 10-90 min with enzyme [2] [8] |
| Success Rate | Generally high; considered the first-line method [29] [28] | Lower and more variable [28] |
| Risk of Tissue Damage | Moderate (overheating can damage morphology) | High (over-digestion can destroy epitopes and architecture) [2] [8] |
| Primary Advantage | Broad applicability and effectiveness | Can be effective for specific, resilient antigens [8] |
The following workflow provides a logical framework for determining the necessity for and selection of an antigen retrieval method, integrating the considerations of tissue type and fixation.
Experimental Data and Protocol Comparison
Quantitative Comparison of Retrieval Methods
Recent research provides quantitative evidence for the differential effectiveness of HIER and PIER, depending on the target antigen and tissue type.
Table 3: Experimental Outcomes from Antigen Retrieval Method Comparisons
| Study Focus | Tissue Type | Target Antigen | Tested Methods | Key Finding | Source |
|---|---|---|---|---|---|
| Glycoprotein Detection | Osteoarthritic Cartilage | CILP-2 | HIER, PIER, HIER+PIER, Control | PIER produced the most abundant CILP-2 staining. HIER alone or combined with PIER was less effective. | [8] |
| Infectious Disease Research | Murine Vaginal Tissue | Eosinophil Protein | HIER (Citrate, 80°C), PIER (Proteinase K) | HIER increased antibody binding visibly and preserved tissue morphology best for automated analysis. | [15] |
Detailed Experimental Protocols
Protocol 1: Standard HIER Using a Pressure Cooker [6] This is a widely used and robust method for HIER.
- Deparaffinize and Rehydrate sections using xylene and graded ethanol series to water.
- Add Buffer: Fill a domestic pressure cooker with an appropriate antigen retrieval buffer (e.g., 10 mM Sodium Citrate, pH 6.0, or Tris-EDTA, pH 9.0).
- Heat: Place the open cooker on a hot plate at full power until the buffer boils.
- Load Slides: Carefully transfer slides to the rack in the boiling buffer.
- Pressurize: Secure the lid. Once full pressure is reached, time for 3 minutes.
- Cool: Turn off heat, place cooker in sink, and run cold water to depressurize. Open lid and run cold water over slides for 10 minutes.
- Proceed: Continue with IHC staining protocol (peroxide block, etc.).
Protocol 2: PIER Using Proteinase K [8] [15] This protocol is adapted from studies on cartilage and murine reproductive tissue.
- Deparaffinize and Rehydrate sections.
- Prepare Enzyme Solution: Create a working solution of Proteinase K (e.g., 30 µg/mL or 0.6 units/mL) in an appropriate buffer (e.g., 50 mM Tris/HCl, pH 6.0).
- Digest: Pipette the solution onto the slides and incubate in a humidity chamber for 15-90 minutes at 37°C. Note: Time is highly antigen-specific and requires optimization.
- Rinse: Gently rinse slides with TBS or PBS.
- Proceed: Continue with the standard IHC staining protocol.
The Scientist's Toolkit: Essential Reagents and Materials
Table 4: Key Research Reagent Solutions for Antigen Retrieval
| Item | Function/Description | Example Uses |
|---|---|---|
| Citrate Buffer (pH 6.0) | A low-pH retrieval buffer for HIER. Effective for a wide range of antigens. | Often the first-choice buffer for optimization matrices [6] [29]. |
| Tris-EDTA Buffer (pH 9.0) | A high-pH retrieval buffer for HIER. Crucial for unmasking many nuclear and phospho-antigens. | Essential for testing when citrate fails; used in CILP-2 study [6] [8]. |
| Proteinase K | A broad-spectrum serine protease used in PIER. Cleaves peptide bonds. | Used for enzymatic retrieval in cartilage and vaginal tissue studies [8] [15]. |
| Sodium Borohydride | Aldehyde quencher. Reduces free aldehyde groups from fixation that cause background. | Particularly useful after glutaraldehyde fixation to reduce non-specific staining [11]. |
| Pressure Cooker / Decloaking Chamber | Device to achieve consistent high-temperature heating for HIER. | Provides rapid, uniform heating; recommended for the standard 3-minute protocol [6]. |
The necessity for antigen retrieval is not a binary question but a strategic decision rooted in tissue preparation. Antigen retrieval is a critical and typically mandatory step for FFPE tissues due to formalin-induced epitope masking, while it is usually unnecessary for frozen tissues where native epitopes are preserved.
Between the two primary methodologies, HIER is the recommended first-line approach due to its broader effectiveness and lower risk of tissue damage compared to PIER. However, as evidenced by recent research, PIER can be superior for specific antigens, such as the cartilage glycoprotein CILP-2 [8]. A systematic optimization of buffer pH, temperature, and incubation time is non-negotiable for achieving specific, reproducible, and high-quality IHC results. The provided protocols and framework offer researchers a clear pathway to determine when retrieval is necessary and how to implement it effectively.
Mastering the Protocols: A Step-by-Step Guide to HIER and PIER Techniques
Heat-Induced Epitope Retrieval (HIER) is a fundamental technique in immunohistochemistry (IHC) that reverses the epitope masking caused by formalin fixation [30]. By heating tissue sections in specific buffer solutions, HIER breaks the methylene bridges formed during fixation, thereby restoring the antibody's ability to bind to its target epitope [2]. The selection of appropriate equipment is crucial for successful and reproducible antigen retrieval, as different heating methods directly impact the temperature uniformity, heating rate, and ultimately, the effectiveness of epitope unmasking [6] [28]. This application note provides a detailed comparison of the most common HIER equipment platforms—microwave, pressure cooker, steamer, and water bath—to guide researchers in selecting the optimal methodology for their specific experimental needs.
Comparative Analysis of HIER Equipment
The choice of heating apparatus significantly influences the conditions of the antigen retrieval process. Below is a systematic comparison of the four primary HIER equipment types.
Table 1: Comparative Analysis of HIER Equipment Choices
| Equipment Type | Typical Temperature Range | Typical Incubation Time | Key Advantages | Principal Limitations |
|---|---|---|---|---|
| Pressure Cooker | 120°C (at full pressure) [6] [2] | 1-5 minutes [6] [2] | Rapid processing; high temperature ensures effective retrieval for many targets [6]. | Potential for tissue detachment; requires careful handling [8] [6]. |
| Microwave | 95-100°C [6] [31] | ~20 minutes once at temperature [6] | Widely accessible; suitable for most antigens [6]. | Risk of "hot spots" and uneven retrieval; buffer evaporation can be an issue [6] [30]. |
| Steamer / Water Bath | 95-100°C [6] | 20 minutes [6] | Gentle heating; minimal risk of tissue detachment; suitable for delicate tissues [6]. | Longer processing times compared to pressure cooking [6]. |
| Water Bath (Low-Temp) | 60-80°C [6] [15] | Overnight [6] | Ideal for tissues prone to detachment (e.g., bone, cartilage, skin) [6]. | Very long incubation time, which may not be suitable for all workflows [6]. |
Detailed HIER Protocols by Equipment Type
Pressure Cooker Protocol
The pressure cooker method utilizes high temperature under pressure to achieve rapid and effective antigen retrieval [6] [2].
- Materials Required: Domestic stainless steel pressure cooker, hot plate, vessel with slide rack, antigen retrieval buffer (e.g., Citrate pH 6.0, Tris-EDTA pH 9.0) [6].
- Step-by-Step Procedure:
- Add antigen retrieval buffer to the pressure cooker and place it on a hot plate at full power [6].
- While the buffer is heating, deparaffinize and rehydrate the tissue sections [6].
- Once the buffer is boiling, transfer the slides from tap water into the pressure cooker. Rest the lid on top but do not secure it yet [6].
- Secure the pressure cooker lid as per the manufacturer's instructions. Once full pressure is reached, time for 3 minutes [6].
- After 3 minutes, turn off the hotplate, place the cooker in a sink, and activate the pressure release valve. Run cold water over the cooker to depressurize [6].
- Open the lid and run cold water over the slides for 10 minutes to cool them and allow the antigenic sites to re-form [6].
- Continue with the standard IHC staining protocol [6].
Scientific Microwave Protocol
A scientific microwave provides temperature control for more consistent results than domestic models [6].
- Materials Required: Scientific microwave (or domestic 850W microwave), microwaveable vessel with slide rack or Coplin jar, antigen retrieval buffer [6].
- Step-by-Step Procedure:
- Deparaffinize and rehydrate the tissue sections [6].
- Place slides in a microwaveable vessel and add sufficient antigen retrieval buffer to cover them by a few centimeters. Use a non-sealed vessel to allow for evaporation [6].
- Place the vessel in the microwave.
- Monitor the buffer level closely during heating to prevent the slides from drying out, adding more buffer if necessary [6].
- After 20 minutes, remove the vessel and run cold tap water into it for 10 minutes to cool the slides [6].
- Proceed with the IHC staining protocol [6].
Vegetable Steamer Protocol
The steamer method provides a gentle, consistent heat at approximately 95-100°C, minimizing the risk of tissue damage [6].
- Materials Required: Vegetable steamer, vessel with slide rack, antigen retrieval buffer [6].
- Step-by-Step Procedure:
- Deparaffinize and rehydrate the tissue sections [6].
- Set up the vegetable steamer and preheat it according to the manufacturer's instructions [6].
- Pre-heat the antigen retrieval buffer to boiling in a separate flask [6].
- Put the container that will hold the rack of slides into the steamer. Carefully add the pre-heated buffer to the container, followed by the rack of slides. Close the lid of the steamer and the container [6].
- Incubate the slides for 20 minutes once the system is stabilized at 95-100°C [6].
- After 20 minutes, remove the vessel and run cold tap water into it for 10 minutes [6].
- Continue with the standard IHC staining protocol [6].
Water Bath Protocol
Water baths are particularly useful for low-temperature, long-term retrieval, which is beneficial for tissues that are prone to detachment [6] [15].
- Materials Required: Water bath, vessel with slide rack, antigen retrieval buffer [6].
- Step-by-Step Procedure:
- Deparaffinize and rehydrate the tissue sections [6].
- Pre-heat the antigen retrieval buffer in the water bath. The temperature can be set to a range from 60°C up to 95-100°C, depending on the protocol and tissue requirements [6] [15].
- Place the slides in the pre-heated buffer.
- For high-temperature retrieval (95-100°C), incubate for 20 minutes [6].
- For low-temperature retrieval (e.g., 80°C), a longer incubation may be used, such as 20 minutes as demonstrated in a study on vaginal tissue [15].
- For very delicate tissues like bone or cartilage, a water bath set to 60°C with an overnight incubation can be used to prevent section detachment [6].
- After incubation, remove the vessel and cool the slides at room temperature or by running cold water for 10 minutes [6].
- Proceed with the IHC staining protocol [6].
The Scientist's Toolkit: Essential Research Reagents & Materials
Successful HIER relies on a set of core reagents and materials. The selection of buffer and its pH is often more critical than the chemical composition itself for effective retrieval [30].
Table 2: Essential Reagents and Materials for HIER
| Item | Function / Description | Examples & Notes |
|---|---|---|
| Antigen Retrieval Buffers | Solution in which slides are heated; pH is critical for success [6] [30]. | Citrate Buffer (pH 6.0): A common, low-pH solution [6] [31]. Tris-EDTA (pH 9.0): A common, high-pH solution [6] [31]. EDTA (pH 8.0): Another high-pH option [6] [31]. |
| Slide Rack and Vessel | Holds slides during the retrieval process. | Must be compatible with the heating method (e.g., metal for pressure cooker, plastic/microwave-safe for microwave) [6]. |
| Adhesive Microscope Slides | Provides strong adhesion for tissue sections during harsh heating steps. | Poly-L-lysine coated, APES coated, or positively charged slides prevent tissue detachment [8] [30]. |
| Heating Apparatus | Equipment used to heat the retrieval buffer. | Pressure cooker, microwave, vegetable steamer, or water bath [6]. |
| Primary Antibody | Binds specifically to the target antigen. | Validation data from the manufacturer should be consulted for recommended retrieval conditions [2]. |
Experimental Workflow and Strategic Selection
The following diagram illustrates the decision-making workflow for selecting and optimizing a HIER protocol, integrating equipment choice with buffer selection.
HIER Equipment and Buffer Selection Workflow
The choice of HIER equipment is a critical determinant in the success of immunohistochemistry, directly impacting epitope retrieval efficiency, tissue morphology preservation, and protocol reproducibility. As demonstrated, each platform—pressure cooker, microwave, steamer, and water bath—offers a distinct set of advantages tailored to different experimental needs, from rapid high-temperature unmasking to gentle retrieval for delicate tissues. A systematic approach to optimization, which includes empirical testing of retrieval buffers and equipment parameters, is essential for developing a robust IHC protocol. By carefully considering the specific antigen, tissue type, and the guidance provided in this application note, researchers can reliably select and implement the most appropriate HIER methodology to advance their research and drug development projects.
Heat-Induced Epitope Retrieval (HIER) has dramatically improved our ability to detect antigens in formalin-fixed, archival tissues by partially reversing or disrupting the aldehyde cross-links formed during fixation [32]. The composition and pH of the retrieval buffer are among the most critical factors influencing HIER efficacy, alongside the amount of heat and duration of heating [32]. Selecting the appropriate retrieval buffer is essential for restoring epitope conformation and enabling accurate antibody binding, which is crucial for reproducible immunohistochemistry (IHC) results in research and diagnostic applications [32] [2]. This guide provides a detailed comparison of the three most prevalent HIER buffers—Citrate (pH 6.0), Tris-EDTA (pH 9.0), and EDTA (pH 8.0)—to inform method optimization within the broader context of HIER vs. PIER research.
Comparative Analysis of Key HIER Buffers
The choice of buffer can determine the success or failure of antigen detection. The table below summarizes the core characteristics of these critical buffers for direct comparison.
Table 1: Key Characteristics of Critical HIER Buffers
| Buffer | Typical pH | Chemical Composition | Primary Mechanism | Antigen Examples |
|---|---|---|---|---|
| Citrate Buffer | 6.0 [32] [6] | 10 mM Sodium Citrate, 0.05% Tween 20 [6] | Hydrolytic cleavage of cross-links [32] | CD5, CD35, BerEP4 [32] |
| Tris-EDTA Buffer | 9.0 [32] [6] | 10 mM Tris, 1 mM EDTA, 0.05% Tween 20 [6] | Calcium ion chelation & thermal unfolding [32] [2] | p27, many nuclear antigens [33] |
| EDTA Buffer | 8.0 [32] [6] | 1 mM EDTA [6] | Calcium ion chelation from coordination complexes [32] | - |
Citrate Buffer (pH 6.0)
- Applications and Performance: Citrate buffer at pH 6.0 is a very popular retrieval medium and is effective for a wide range of antigens, including many lymphocyte subset antigens and oncoproteins [32]. It can be used to retrieve epitopes that are not otherwise detectable in formalin-fixed, paraffin-wax sections and can sometimes substitute for enzymatic digestion [32].
- Advantages and Limitations: Its primary advantage is its widespread success and relatively gentle effect on tissue morphology compared to EDTA-containing solutions [32]. It serves as an excellent starting point for optimization.
Tris-EDTA Buffer (pH 9.0)
- Applications and Performance: This high-pH buffer is particularly effective for many nuclear antigens, transcription factors, and phospho-proteins. For instance, detection of p27 is significantly enhanced following HIER in basic (pH 9.0-9.5) solutions compared to no treatment or acidic buffers [33].
- Advantages and Limitations: The alkaline environment combined with the chelating action of EDTA often provides robust antigen recovery for challenging targets. However, tissues treated with this or other EDTA-containing solutions may show enhanced tissue damage compared to citrate-based retrieval buffers [32].
EDTA Buffer (pH 8.0)
- Applications and Performance: EDTA-based solutions provide excellent antigen recovery [32]. The mechanism is hypothesized to involve the chelation of calcium ions from coordination complexes with proteins, thereby breaking cross-links [32] [2].
- Advantages and Limitations: While highly effective, its use may be associated with more prominent tissue deterioration, requiring careful validation of incubation times [32].
Detailed Experimental Protocols
Standardized HIER Protocol for Buffer Comparison
A consistent HIER methodology is essential for fairly evaluating different buffers. The following protocol can be applied using a pressure cooker, microwave, or steamer.
Materials Required:
- Deparaffinized and rehydrated tissue sections
- Antigen retrieval buffer (Citrate pH 6.0, Tris-EDTA pH 9.0, or EDTA pH 8.0)
- Heating device (pressure cooker, scientific microwave, or vegetable steamer)
- Slide rack and Coplin jar or suitable container
- Hot plate (if using a pressure cooker)
Procedure:
- Preparation: Add a sufficient volume of antigen retrieval buffer to cover slides by at least a few centimeters in a suitable container [6].
- Heating (Pressure Cooker Method):
- Place the container with buffer on a hot plate and bring to a boil [6].
- Carefully transfer slides into the boiling buffer, secure the lid, and allow the cooker to reach full pressure [6].
- As soon as full pressure is reached, time for 3 minutes [6].
- After 3 minutes, turn off the heat, depressurize, and run cold water over the cooker for 10 minutes to cool the slides [6].
- Heating (Microwave Method):
- Place slides in buffer within a microwaveable vessel and heat in a scientific microwave at 98°C for 20 minutes [6]. If using a domestic microwave, boil at full power for 20 minutes, monitoring for evaporation.
- After heating, remove the vessel and run cold tap water into it for 10 minutes to cool [6].
- Completion: Once cooled, proceed with the standard IHC staining protocol (blocking, primary antibody incubation, detection, etc.) [6].
Protocol Optimization and Validation
Optimal retrieval conditions are influenced by the tissue type, fixation method, and target antigen, necessitating systematic optimization [34] [33].
- Systematic Optimization Matrix: For a new antibody, create a testing matrix that evaluates different combinations of buffer pH and heating time [34]. This empirical approach is the most reliable way to determine the optimal conditions for a specific antigen-antibody pair.
- Essential Controls:
- No-Retrieval Control: A section processed without any HIER treatment helps determine if the retrieval itself is necessary or introduces artifacts [34].
- Positive Control: A tissue with known expression of the target antigen confirms that the protocol and reagents are working correctly [2].
- Negative Control: A section processed without the primary antibody checks for non-specific binding from the detection system [2].
The following workflow diagrams a systematic approach for optimizing HIER conditions, integrating the key decision points and validation steps discussed.
The Scientist's Toolkit: Essential Research Reagents
A successful HIER workflow relies on several key reagents and equipment. The following table lists these essential items and their functions.
Table 2: Essential Reagents and Equipment for HIER Protocols
| Item | Function / Description | Examples / Notes |
|---|---|---|
| Citrate Buffer | Acidic retrieval solution; hydrolyzes cross-links [32]. | 10 mM Sodium citrate, 0.05% Tween 20, pH 6.0 [6]. |
| Tris-EDTA Buffer | Alkaline retrieval solution; chelates ions & unfolds protein [32] [2]. | 10 mM Tris, 1 mM EDTA, 0.05% Tween 20, pH 9.0 [6]. |
| EDTA Buffer | Chelating retrieval solution; extracts calcium ions [32]. | 1 mM EDTA, pH 8.0 [6]. |
| Heating Device | Applies heat to disrupt protein cross-links. | Pressure cooker, scientific microwave, steamer, or water bath [32] [6]. |
| Proteolytic Enzymes | For PIER; enzymatically degrade cross-links [2]. | Trypsin, Proteinase K, Pepsin [2] [35]. |
| Validated Antibodies | Primary antibodies with known performance in IHC. | Check manufacturer's datasheet for recommended retrieval method [2]. |
The strategic selection and optimization of HIER buffers—Citrate (pH 6.0), Tris-EDTA (pH 9.0), and EDTA (pH 8.0)—are foundational to successful antigen detection in formalin-fixed tissues. There is no universal "best" buffer; the optimal choice is antigen-dependent [32] [6]. A systematic empirical approach, starting with a pH matrix and rigorous validation using appropriate controls, is the most reliable path to a robust, reproducible IHC protocol. This rigorous methodology ensures that HIER continues to be an indispensable tool for researchers and drug development professionals, enabling the accurate visualization of biomarkers critical for diagnostic and therapeutic discovery.
In immunohistochemistry (IHC), antigen retrieval is a critical step for reversing the protein cross-linking caused by formalin fixation, which masks antigenic sites and hinders antibody binding [36]. The pH of the retrieval solution used in Heat-Induced Epitope Retrieval (HIER) significantly influences staining outcomes by altering protein conformation and electrostatic charges, thereby affecting antibody-epitope binding efficiency [37]. Understanding how different antigens respond to pH variations is fundamental to developing reliable IHC protocols, especially when choosing between HIER and Proteolytic-Induced Epitope Retrieval (PIER) methods [36] [7].
Research indicates that antigens demonstrate distinct, predictable profiles in response to the pH of the retrieval buffer [37]. These profiles have been systematically categorized into four main types: Stable, V-type, Increasing, and Decreasing. This classification provides a strategic framework for researchers to optimize antigen retrieval conditions, thereby enhancing staining intensity, specificity, and overall reproducibility for a wide range of targets [37].
Classification of Antigen pH Profiles
The response of antigens to the pH of heat-induced retrieval buffers can be broadly classified into four distinct patterns. Understanding these profiles allows for systematic optimization of immunohistochemistry protocols. The table below summarizes the key characteristics of each profile.
Table 1: Classification of Antigen pH Response Profiles
| Profile Type | Description of Staining Response | Representative Antigen Examples |
|---|---|---|
| Stable Type | pH has minimal to no significant effect on staining results. | PCNA, AE1, EMA, CD20 [37] |
| V-type | Both high and low pH values yield good staining, while intermediate pH (e.g., 4-5) results in poorer staining. | Estrogen Receptor (ER), Ki-67 [37] |
| Increasing Type | Staining results progressively improve with increasing pH. | HMB45 [37] |
| Decreasing Type | Staining results weaken as the pH increases. | MOC31 [37] |
Stable Type Profile
For antigens with a Stable Type profile, the staining intensity and quality remain consistent across a wide pH spectrum [37]. This characteristic simplifies protocol development, as the choice of retrieval buffer pH is less critical. Antigens such as Proliferating Cell Nuclear Antigen (PCNA) and epithelial markers like EMA and AE1 fall into this category. For these targets, researchers can prioritize buffer choice based on other factors, such as tissue preservation or compatibility with other steps in the IHC workflow.
V-type Profile
The V-type profile is characterized by optimal staining at both acidic (low) and basic (high) pH conditions, with a notable drop in staining quality at neutral or slightly acidic pH ranges (around pH 4-5) [37]. This biphasic response suggests that the epitope may adopt different conformations that are favorably exposed at pH extremes but masked at intermediate pH levels. Key nuclear markers like Ki-67 and the Estrogen Receptor (ER) exhibit this behavior, requiring careful selection of retrieval buffer to avoid the suboptimal middle pH range.
Increasing Type Profile
Antigens with an Increasing Type profile show a direct correlation between retrieval buffer pH and staining intensity [37]. The higher the pH, the more robust the staining result. The melanoma marker HMB45 is a documented example of this profile. For such targets, high-pH buffers like Tris-EDTA (pH 8.0-9.0) are typically the most effective choice and should be the starting point for protocol optimization.
Decreasing Type Profile
The Decreasing Type profile is the inverse of the Increasing Type, where staining intensity diminishes as the pH of the retrieval buffer increases [37]. This pattern is considered rare, with antibodies like MOC31 serving as examples. For these antigens, low-pH buffers, such as sodium citrate (pH 6.0), are more likely to yield successful results.
Experimental Protocols for pH Profile Determination
Determining the pH profile of a novel or uncharacterized antigen requires a systematic experimental approach. The following protocol outlines a standard method for mapping antigen response to pH using Heat-Induced Epitope Retrieval (HIER).
Materials and Reagents
Table 2: Essential Research Reagent Solutions for Antigen Retrieval Optimization
| Reagent / Material | Function / Purpose | Example Specifications / Notes |
|---|---|---|
| Sodium Citrate Buffer | Low-pH retrieval solution (e.g., pH 6.0) [6] | 10 mM Tri-sodium citrate, 0.05% Tween 20 [6] |
| Tris-EDTA Buffer | High-pH retrieval solution (e.g., pH 9.0) [6] | 10 mM Tris base, 1 mM EDTA, 0.05% Tween 20 [6] |
| EDTA Buffer | High-pH retrieval solution (pH 8.0-9.0) [37] | 1 mM EDTA, pH 8.0 [6] |
| Proteinase K | Enzyme for Proteolytic-Induced Epitope Retrieval (PIER) [36] | Working concentration: 20 µg/mL; Incubation: 37°C for 20 min [36] |
| Trypsin | Enzyme for Proteolytic-Induced Epitope Retrieval (PIER) [36] | Working concentration: 0.05% to 0.1%; Incubation: 37°C for 10-40 min [36] |
| Pepsin | Enzyme for Proteolytic-Induced Epitope Retrieval (PIER) [36] | Working concentration: 0.4%; Incubation: 37°C for 30-180 min [36] |
| Adhesive Microscope Slides | Prevents tissue detachment during rigorous HIER treatments [17] | Essential for challenging tissues like cartilage [17] |
Step-by-Step HIER pH Optimization Protocol
The following workflow illustrates the key stages in the experimental process for determining antigen pH profiles.
Diagram 1: Experimental workflow for determining antigen pH profiles using HIER.
Section Preparation: Cut 4 µm thick sections from Formalin-Fixed, Paraffin-Embedded (FFPE) tissue blocks and mount them on adhesive microscope slides to prevent detachment [17]. Deparaffinize and rehydrate the sections using xylene and a graded ethanol series [17].
Buffer Selection and Heating: Prepare a set of antigen retrieval buffers covering acidic, neutral, and basic pH ranges. Common choices include:
- Acidic: Sodium citrate buffer (10 mM, pH 6.0) [6].
- Neutral: PBS buffer (pH 7.0) [36].
- Basic: Tris-EDTA (pH 9.0) or EDTA (pH 8.0) [6]. Immerse slides in the preheated retrieval buffer and perform HIER using a validated heating apparatus (microwave, pressure cooker, or steamer). A common starting condition is 95°C for 20 minutes [6] [7].
Cooling and Staining: After heating, remove the container from the heat source and cool the slides by running cold tap water into it for 10-15 minutes. This gradual cooling helps the antigenic sites re-form into their stable configurations [6]. Proceed with the standard IHC staining protocol, including blocking, primary antibody incubation, and detection.
Analysis and Profiling: Analyze the stained slides microscopically. Compare the staining intensity, signal-to-noise ratio, and cellular localization across the different pH conditions. Classify the antigen's behavior according to the four primary profiles (Stable, V-type, Increasing, Decreasing) based on the results.
Experimental Design Matrix
To efficiently optimize conditions, a multi-slide experimental matrix is recommended. The table below outlines a standard setup for investigating the effects of pH and retrieval time simultaneously.
Table 3: Experimental Matrix for Optimizing Retrieval Time and Buffer pH
| Incubation Time | Antigen Retrieval Solution pH | ||
|---|---|---|---|
| Acidic (e.g., pH 6.0) | Neutral (e.g., pH 7.0) | Basic (e.g., pH 9.0) | |
| 4 minutes | Slide #1 | Slide #2 | Slide #3 |
| 8 minutes | Slide #4 | Slide #5 | Slide #6 |
| 12 minutes | Slide #7 | Slide #8 | Slide #9 |
Adapted from optimization protocols provided by Boster Bio [37].
Integration with Broader Antigen Retrieval Strategies
While pH optimization is a powerful tool, it is one component of a comprehensive antigen retrieval strategy. The choice between HIER and PIER remains antigen-dependent.
HIER vs. PIER in Context
Heat-Induced Epitope Retrieval (HIER) is generally the first-line method due to its gentler effect on tissue morphology and higher success rate for a broad range of antigens [36] [38]. It offers more controllable parameters, primarily through the adjustment of buffer pH, temperature, and heating time [36]. Proteolytic-Induced Epitope Retrieval (PIER), using enzymes like proteinase K, trypsin, or pepsin, can be effective for difficult-to-recover epitopes that do not respond well to heat [36] [28]. However, PIER carries a higher risk of damaging tissue morphology and destroying the antigen itself if not meticulously optimized [28].
Case Study: The Critical Role of Tissue Context
A recent study on osteoarthritic cartilage highlights that the optimal retrieval method can be highly specific to the tissue and target protein. For detecting the cartilage glycoprotein CILP-2, PIER (using proteinase K and hyaluronidase) produced superior staining compared to HIER alone [17] [8]. Furthermore, combining HIER with PIER did not improve results and often led to tissue detachment, underscoring the need for empirical testing in challenging matrices like cartilage [17]. This case demonstrates that while pH profiling for HIER is a vital guide, it must be applied within the context of the specific tissue and antigen system.
The systematic classification of antigens into Stable, V-type, Increasing, and Decreasing pH profiles provides a critical framework for optimizing IHC assays. By understanding and applying these profiles, researchers can move beyond trial-and-error and make informed decisions about buffer selection for HIER. This approach significantly enhances the reliability, intensity, and specificity of immunohistochemical staining. For targets unresponsive to HIER optimization, alternative methods like PIER should be investigated, always considering the unique context of the target tissue and antigen.
In the broader context of optimizing antigen retrieval methods for immunohistochemistry (IHC), researchers must navigate the critical choice between Heat-Induced Epitope Retrieval (HIER) and Proteolytic-Induced Epitope Retrieval (PIER). While HIER has become the more common approach due to its gentler nature on tissue morphology, PIER remains an indispensable tool for specific applications, particularly for difficult-to-retrieve epitopes and certain tissue types [2] [19]. PIER employs proteolytic enzymes such as trypsin, proteinase K, and pepsin to cleave protein crosslinks formed during formalin fixation, thereby restoring antigenic accessibility that enables antibody binding [2] [39]. This application note provides detailed protocols and optimization strategies for implementing PIER methods effectively within a research setting, with specific consideration for drug development applications where precise protein localization and quantification are paramount.
The fundamental challenge addressed by all antigen retrieval methods stems from formalin fixation, which creates methylene bridges between proteins, thereby altering protein structure and masking epitopes from antibody recognition [2]. Whereas HIER utilizes heat to disrupt these crosslinks, PIER works through enzymatic degradation of the proteins surrounding the epitope [40]. Each enzyme has specific cleavage characteristics and optimal working conditions that must be carefully matched to the target antigen and tissue type to achieve optimal results while preserving tissue integrity.
Enzyme Selection and Comparative Analysis
The selection of an appropriate enzyme is the foundational step in developing a successful PIER protocol. The three most commonly used enzymes—trypsin, proteinase K, and pepsin—each possess distinct characteristics, making them suitable for different applications. The table below provides a systematic comparison of their optimal working conditions:
Table 1: Optimal Working Conditions for Common PIER Enzymes
| Enzyme | Typical Concentration | Incubation Conditions | Buffer Solution | pH | Primary Applications |
|---|---|---|---|---|---|
| Trypsin | 0.05% solution [41] | 10-20 minutes at 37°C [41] [42] | 0.05% trypsin, 1% CaCl₂, adjusted with NaOH [41] | 7.8 [41] | General purpose protein digestion; often recommended for cytoplasmic and membrane antigens [2] |
| Proteinase K | 20 μg/mL (1x) [41] | 10-20 minutes at 37°C [41]; Extended incubations (90 minutes) possible with specific protocols [17] | TE Buffer (Tris-EDTA), pH 8.0 [41] | 8.0 [41] | Difficult epitope retrieval; often combined with hyaluronidase for dense extracellular matrices like cartilage [17] |
| Pepsin | Not specified in results | 10-20 minutes at 37°C [42] | Not specified in results | Not specified in results | Particularly useful for cytokeratins and immunoglobulins [40] |
The selection criteria should extend beyond these basic parameters to include the specific epitope characteristics, tissue type, and degree of fixation. Trypsin generally offers a balanced approach for many applications, while proteinase K provides a more aggressive retrieval beneficial for heavily crosslinked or masked epitopes [2]. Pepsin has demonstrated particular value for certain antigen classes, including cytokeratins and immunoglobulins [40].
Detailed Experimental Protocols
Standardized PIER Workflow for IHC
The diagram below illustrates the generalized workflow for incorporating PIER into a standard IHC protocol, highlighting key decision points and quality control measures:
Enzyme-Specific Protocol Formulations
Trypsin Digestion Protocol
Working Solution Preparation:
- Combine 1 ml of 0.5% trypsin stock solution with 1 ml of 1% calcium chloride stock solution [41]
- Add 8 ml of distilled water for a total volume of 10 ml [41]
- Adjust pH to 7.8 with 1N NaOH [41]
Method:
- Apply the trypsin working solution to cover tissue sections completely
- Transfer slides to a humidified chamber and incubate for 10-20 minutes at 37°C [41] [42]
- Remove slides from incubator and allow to cool at room temperature for 10 minutes [41]
- Rinse in running tap water for 3 minutes to terminate enzymatic activity [42]
- Proceed with standard IHC staining protocol
Proteinase K Digestion Protocol
Working Solution Preparation:
- Prepare 1x proteinase K working solution (20 μg/ml) by diluting 20X stock solution in TE buffer, pH 8.0 [41]
- Mix well to ensure homogeneous distribution of the enzyme
Method:
- Cover tissue sections completely with proteinase K working solution
- Incubate for 10-20 minutes at 37°C in a humidified chamber [41]
- For challenging tissues or epitopes, extended incubation times (up to 90 minutes) may be required [17]
- Allow sections to cool at room temperature for 10 minutes [41]
- Rinse gently with distilled water or appropriate buffer
- Continue with standard IHC staining procedure
Enhanced Protocol for Challenging Tissues
Research on decalcified joint tissues and cartilage matrix proteins has demonstrated that certain challenging specimens may require modified PIER approaches:
Combined Enzymatic Retrieval for Cartilage Tissues:
- Apply Proteinase K solution (30 μg/mL in 50 mM Tris/HCl, 5 mM CaCl₂ solution, pH 6.0) for 90 minutes at 37°C [17]
- Subsequently treat with 0.4% bovine hyaluronidase in HEPES-buffered medium for 3 hours at 37°C [17]
- This combined approach has proven effective for retrieving difficult epitopes within dense extracellular matrices, such as cartilage intermediate layer protein 2 (CILP-2) [17]
Research Reagent Solutions
Successful implementation of PIER requires specific reagent systems optimized for enzymatic epitope retrieval. The following table outlines essential solutions and their functions:
Table 2: Essential Research Reagents for PIER Protocols
| Reagent Solution | Composition | Primary Function | Application Notes |
|---|---|---|---|
| Trypsin Working Solution | 0.05% trypsin, 1% CaCl₂, pH 7.8 [41] | Proteolytic digestion of protein crosslinks | Calcium chloride stabilizes enzyme activity; optimal for general purpose antigen retrieval |
| Proteinase K Solution | 20 μg/mL in TE Buffer, pH 8.0 [41] | Broad-spectrum proteolysis for difficult epitopes | Effective for dense tissue matrices; can be combined with hyaluronidase for enhanced retrieval [17] |
| TE Buffer | Tris-EDTA, pH 8.0 [41] | Optimal buffer for Proteinase K | Maintains enzymatic activity while preserving tissue architecture |
| Enzyme Diluent Buffer | 50 mM Tris/HCl, 5 mM CaCl₂, pH 6.0 [17] | Stabilizing medium for extended enzymatic retrieval | Used in enhanced protocols for challenging tissues like cartilage |
| Hyaluronidase Solution | 0.4% in HEPES-buffered medium [17] | Digestion of hyaluronic acid in extracellular matrix | Enhances antibody penetration in matrix-rich tissues when combined with Proteinase K |
Troubleshooting and Quality Control
Common Technical Challenges and Solutions
Even with optimized protocols, researchers may encounter technical challenges requiring systematic troubleshooting:
- Tissue Damage or Morphological Alterations: Caused by excessive enzymatic digestion [2]. Solution: Reduce incubation time or enzyme concentration, and ensure precise temperature control at 37°C [2] [19]
- Weak or No Staining: Results from under-retrieval where epitopes remain masked [2]. Solution: Gradually increase incubation time in 2-5 minute increments or consider switching to a more aggressive enzyme (e.g., from trypsin to proteinase K)
- High Background Staining: Caused by over-digestion creating non-specific binding sites [2]. Solution: Reduce enzyme concentration or incubation time, and ensure thorough rinsing after retrieval
- Tissue Detachment from Slides: Particularly problematic with loose-textured or decalcified tissues [14]. Solution: Use poly-L-lysine coated adhesive slides and avoid excessive agitation during rinsing steps
Essential Quality Control Measures
Robust experimental design requires implementation of comprehensive controls to ensure specificity and reproducibility:
- Negative Controls: Sections processed without primary antibody to identify non-specific binding from the secondary antibody system [2]
- Positive Controls: Tissues with known expression of the target antigen to confirm protocol effectiveness [2]
- Specificity Controls: Whenever possible, employ knockout/knockdown validation or blocking peptides to confirm target specificity [2]
- Morphological Assessment: Always include a section stained with standard H&E to evaluate tissue preservation after PIER treatment
Within the broader context of antigen retrieval optimization, PIER represents a specialized but invaluable approach for specific research scenarios. While HIER generally serves as the initial method of choice due to its gentler impact on tissue morphology and more definable parameters [2] [7], PIER demonstrates particular strength for difficult-to-retrieve epitopes, heavily crosslinked tissues, and specific antigen classes such as cytokeratins and immunoglobulins [40]. Recent research on challenging tissues like decalcified joint structures and cartilage matrix proteins has reinforced PIER's unique value, with studies showing superior performance compared to HIER for certain targets like CILP-2 [17].
The decision framework for implementing PIER should consider both the target antigen characteristics and tissue preservation requirements. PIER is particularly indicated when: (1) HIER methods produce suboptimal staining despite buffer and pH optimization; (2) working with antigens known to be susceptible to heat degradation; (3) processing tissues with dense extracellular matrices that limit antibody penetration; and (4) when targeting specific protein classes known to respond well to enzymatic retrieval. By understanding the precise applications, optimal conditions, and limitations of each enzymatic approach, researchers can strategically incorporate PIER into their IHC optimization pipeline, thereby expanding the range of detectable targets and enhancing the quality of protein localization data in both basic research and drug development applications.
Within immunohistochemistry (IHC), the process of formalin fixation creates methylene bridges that cross-link proteins, thereby masking epitopes and restricting antibody access [43] [18]. Heat-Induced Epitope Retrieval (HIER) is a critical technique for reversing this masking, significantly enhancing the detection of target proteins in formalin-fixed, paraffin-embedded (FFPE) tissues [18] [28]. The core principle of HIER involves the application of heat, typically in a buffered solution, to break the formaldehyde-induced cross-links, thereby restoring antigenicity [43]. This protocol focuses on two common HIER methods: the pressure cooker and the microwave. The pressure cooker operates at high temperatures (110-120°C) for short durations, while the microwave method uses lower temperatures (around 98-100°C) for longer periods [43] [6]. Selecting the appropriate method and optimizing the protocol is a fundamental aspect of research comparing HIER with Proteolytic-Induced Epitope Retrieval (PIER), as the efficacy of antigen retrieval can vary dramatically depending on the specific antigen, tissue type, and fixation conditions [8] [28].
Principles and Mechanisms of HIER
The efficacy of HIER is believed to stem from its ability to reverse the chemical modifications imposed by formaldehyde fixation [18]. While the precise mechanism remains an area of investigation, two prominent theories exist. The first posits that the thermal energy provided breaks the peptide cross-links that bind surrounding proteins to the antigen of interest, effectively "unmasking" the epitope [18]. A second theory suggests that HIER, particularly when using certain buffers, acts by chelating bound calcium ions from the sites of cross-links, facilitating their disruption [18]. The process causes crosslinked proteins to unfold, making the hidden epitopes accessible once again for antibody binding [43]. The success of HIER is not universal; some antigens, particularly those in tissues with a dense extracellular matrix like cartilage, may respond better to enzymatic retrieval (PIER) or a combination of methods, highlighting the necessity for empirical optimization in any systematic comparison of antigen retrieval techniques [8].
Essential Reagents and Equipment
Research Reagent Solutions
The choice of retrieval buffer is a critical variable in HIER optimization. The pH of the buffer solution is often more important than its chemical composition, with different antigens showing optimal retrieval at specific pH levels [18] [28].
Table 1: Key Buffers for Heat-Induced Epitope Retrieval (HIER)
| Buffer Name | Composition | pH | Primary Applications and Notes | Source |
|---|---|---|---|---|
| Sodium Citrate Buffer | 10 mM Sodium Citrate, 0.05% Tween 20 | 6.0 | A common, general-purpose buffer suitable for a wide range of antigens. | [6] [44] |
| Tris-EDTA Buffer | 10 mM Tris Base, 1 mM EDTA, 0.05% Tween 20 | 9.0 | Effective for many antigens; alkaline pH is often optimal. EDTA is a calcium chelator. | [18] [6] |
| EDTA Buffer | 1 mM EDTA, 0.05% Tween 20 | 8.0 | Particularly effective for over-fixed specimens and difficult-to-retrieve antigens. Can cause high background. | [6] [44] |
Required Equipment
The selection of heating apparatus directly influences the time and temperature parameters of the protocol. The main options are:
- Pressure Cooker: A domestic stainless steel pressure cooker is sufficient [6]. It provides rapid, even heating and reaches the highest temperatures (110-120°C), making it highly effective for many antigens [18].
- Microwave: Either a domestic microwave (850 W) or a scientific microwave can be used [6]. A scientific microwave is preferred for its temperature control, reducing the risk of uneven retrieval and tissue detachment caused by violent boiling [18] [6].
- Alternative Equipment: Vegetable steamers and water baths (set at 95-100°C) are also viable, typically requiring incubation times of 20-40 minutes [18] [44].
- General Supplies: Slide rack (metal for pressure cooker, plastic for microwave), a vessel to hold 400-500 mL of buffer, a hotplate (for pressure cooker), and forceps [6].
Detailed Experimental Protocols
HIER Using a Pressure Cooker
The pressure cooker method is highly effective due to the high temperatures achieved, which allow for shorter retrieval times [18].
Materials:
- Domestic stainless steel pressure cooker [6]
- Hot plate [6]
- Antigen retrieval buffer (e.g., Citrate pH 6.0 or Tris-EDTA pH 9.0) [6]
- Metal slide rack [6]
Method:
- Add a sufficient volume of antigen retrieval buffer to the pressure cooker to cover the slides by at least a few centimeters [6].
- Place the open pressure cooker on a hot plate set to full power and bring the buffer to a boil [6].
- While waiting, deparaffinize and rehydrate the tissue sections using standard histological techniques [6].
- Once the buffer is boiling, carefully transfer the slides from tap water into the buffer in the pressure cooker using forceps [6].
- Secure the lid of the pressure cooker as per the manufacturer's instructions [6].
- As soon as the cooker reaches full pressure, time 3 minutes [6].
- After 3 minutes, turn off the hotplate and transfer the pressure cooker to an empty sink. Activate the pressure release valve and run cold water over the cooker to depressurize and cool it [6].
- Once depressurized, open the lid and run cold tap water into the cooker for 10 minutes to cool the slides completely [6].
- Proceed with the subsequent steps of your IHC staining protocol [6].
HIER Using a Microwave
The microwave method is accessible but requires careful monitoring to prevent buffer evaporation and uneven heating [18] [6].
Materials:
- Scientific microwave (recommended) or domestic microwave (850 W) [6]
- Microwaveable vessel with a plastic slide rack [6]
- Antigen retrieval buffer [6]
Method:
- Deparaffinize and rehydrate the tissue sections [6].
- Place the slides in a microwaveable vessel and add enough antigen retrieval buffer to cover them by several centimeters [6]. Use a non-sealed vessel to allow for evaporation [6].
- Place the vessel inside the microwave.
- If using a domestic microwave, set it to full power and wait for the solution to boil. Once boiling, continue to boil for 20 minutes, monitoring closely to ensure the slides do not dry out and adding distilled water if necessary [6]. If using a scientific microwave, program it to maintain a temperature of 98°C for 20 minutes [6].
- After 20 minutes, remove the vessel and run cold tap water into it for 10 minutes to cool the slides [6].
- Continue with the standard IHC staining procedure [6].
Method Comparison and Data Presentation
Choosing between a pressure cooker and a microwave involves balancing efficiency, reproducibility, and practicality. The following table and diagram summarize the key differences.
Table 2: Quantitative Comparison of Pressure Cooker vs. Microwave HIER Methods
| Parameter | Pressure Cooker | Microwave |
|---|---|---|
| Typical Temperature | 110°C - 120°C [43] [18] | 98°C - 100°C [6] |
| Typical Incubation Time | 1 - 5 minutes at temperature [43] [28] | 20 minutes at temperature [6] |
| Heat Distribution | Even and consistent [18] | Often uneven, creating hot/cold spots [18] |
| Primary Advantages | Short time, high sensitivity, even heat [18] | Inexpensive, easy to use, rapid heating [18] |
| Primary Disadvantages | Can cause tissue artifacts and damage [18] | Uneven retrieval, tissue detachment, buffer evaporation [18] [6] |
Integration in HIER vs. PIER Research
In the broader context of optimizing antigen retrieval methods, HIER using a pressure cooker or microwave must be systematically compared to Proteolytic-Induced Epitope Retrieval (PIER). PIER relies on enzymes like proteinase K, trypsin, or pepsin to digest protein cross-links [43] [28]. A critical comparative study on cartilage matrix glycoproteins found that PIER alone provided superior staining for the CILP-2 antigen compared to HIER or a combination of HIER and PIER [8]. In that specific experimental setting, the application of heat even reduced the positive effect of enzymatic retrieval and led to frequent section detachment [8]. This underscores that while HIER is highly effective for a broad range of antigens [28], PIER can be the optimal choice for specific targets, particularly in challenging tissues. Therefore, a rigorous research framework should include direct comparison of both techniques, using appropriate controls and quantitative assessment of staining intensity and tissue morphology preservation [8] [28].
Troubleshooting and Optimization
Optimizing HIER is essential for achieving specific and robust staining. Key parameters to optimize include the retrieval buffer pH, heating time, and temperature [28]. A systematic approach involves testing a matrix of different conditions, for example, using various buffer pH levels (acidic, neutral, basic) at multiple time points (e.g., 1, 5, 10 minutes) and comparing the results to a no-retrieval control [28]. If staining remains weak with one method, switching from a microwave to a pressure cooker for higher temperature retrieval, or trying an EDTA-based buffer for a difficult antigen, is recommended [18] [44]. Always include control tissues known to express the target antigen to validate the retrieval protocol and ensure the specificity of antibody binding [28].
Within the broader research on optimizing antigen retrieval methods, the choice between Heat-Induced Epitope Retrieval (HIER) and Proteolytic-Induced Epitope Retrieval (PIER) is a critical determinant of immunohistochemistry (IHC) success. While HIER is often the first-line method due to its gentler profile, PIER remains an essential technique for retrieving epitopes that are resistant to heat-based methods or are situated within dense tissue matrices [7] [45]. This protocol focuses on PIER using two common enzymes, Trypsin and Proteinase K, providing a standardized and detailed methodology for researchers and drug development professionals. The necessity for antigen retrieval arises from the protein cross-linking caused by formalin fixation, which masks antigenic sites and hinders antibody binding [46] [45]. For certain antigens, particularly those in challenging tissues like cartilage or for specific epigenetic markers, PIER has been demonstrated to provide superior results compared to HIER [24] [47]. The following sections provide a comprehensive guide to executing and optimizing PIER, complete with quantitative data comparisons and essential reagent solutions.
Principle of the PIER Method
Proteolytic-Induced Epitope Retrieval (PIER) functions through the enzymatic cleavage of peptides and protein cross-links that are formed during formaldehyde fixation. These cross-links, primarily methylene bridges, physically mask the epitope of interest, preventing antibody access [46] [45]. Enzymes such as Trypsin and Proteinase K degrade these cross-links and surrounding proteins, thereby unmasking the epitope and restoring antibody binding capacity [28]. In contrast, Heat-Induced Epitope Retrieval (HIER) primarily relies on heat to break the cross-links and unfold proteins [46] [30]. A key disadvantage of PIER is the potential for over-digestion, which can damage both the tissue morphology and the antigen itself, making optimization of time and concentration critical [28]. Furthermore, while the pH for PIER incubation is typically neutral (pH 7.4) and less variable than in HIER, the method is generally considered harsher on tissues [7].
The following workflow diagram illustrates the key decision points and steps in the PIER protocol.
Comparative Analysis of Antigen Retrieval Methods
The choice between PIER and HIER is antigen- and tissue-dependent. The following table summarizes the core characteristics of each method to guide initial experimental design.
Table 1: Comparison of Heat-Induced (HIER) and Proteolytic-Induced (PIER) Epitope Retrieval Methods
| Parameter | Heat-Induced Epitope Retrieval (HIER) | Proteolytic-Induced Epitope Retrieval (PIER) |
|---|---|---|
| Fundamental Principle | Uses heat to break protein cross-links and unfold epitopes [46]. | Uses enzymes to digest proteins and cleave cross-links masking the epitope [28]. |
| Common Agents | Citrate buffer (pH 6.0), Tris-EDTA (pH 9.0), EDTA (pH 8.0) [7] [6]. | Trypsin, Proteinase K, Pepsin [7] [45]. |
| Typical Temperature | ~95°C [7] | 37°C [7] |
| Typical Incubation Time | 10-20 minutes [7] | 5-30 minutes (Trypsin: 10-40 min; Proteinase K: ~20 min) [7] [45] |
| Key Advantages | Gentler on tissue morphology; more definable and reproducible parameters [7]. | Useful for epitopes difficult to retrieve with heat; protocol is generally faster [7] [45]. |
| Key Limitations / Risks | Requires optimization of buffer pH; can cause tissue detachment from slides [46] [7]. | Harsher method; excessive digestion can damage tissue morphology and the antigen itself [7] [28]. |
Recent comparative studies underscore the context-dependent efficacy of PIER. For instance, in the immunohistochemical analysis of the cartilage matrix glycoprotein CILP-2, PIER using Proteinase K and hyaluronidase yielded superior staining results compared to HIER or a combination of both methods [24]. Conversely, a study on murine female reproductive tract tissue found that heat-based retrieval in citrate buffer provided better antibody binding and tissue morphology for specific targets than Proteinase K PIER [15]. Furthermore, research on detecting epigenetic DNA modifications (5-mC and 5-hmC) demonstrated a gradient of detection levels, with a Pepsin/HCl-based retrieval method outperforming high-pH Tris-EDTA HIER, which in turn was more effective than low-pH Citrate HIER [47]. These findings highlight the critical need for empirical optimization.
Materials and Reagent Solutions
The Scientist's Toolkit: Essential Reagents for PIER
Table 2: Key Research Reagent Solutions for PIER
| Reagent | Function / Description | Example Specification / Note |
|---|---|---|
| Trypsin | Proteolytic enzyme that cleaves peptide bonds. | Working Concentration: 0.05% to 0.1% [45]. Must be prepared fresh with pH adjusted to 7.6 [45]. |
| Proteinase K | A broad-spectrum serine protease used for protein digestion. | Working Concentration: 20 µg/mL [45]. |
| Pepsin | An enzyme that breaks down proteins, often used in acidic conditions. | Working Concentration: 0.4% [45]. |
| Phosphate-Buffered Saline (PBS) or Tris-Buffered Saline (TBS) | Used for rinsing and preparing enzyme solutions; provides a stable, neutral pH environment. | Typically used at pH 7.4 for enzymatic digestion [7]. |
| Proteinase K Buffer | The buffer supplied with the enzyme to ensure optimal activity. | Follow manufacturer's instructions; often contains Tris-HCl, CaCl₂, etc. |
| Superfrost Plus Microscope Slides | Charged or coated slides to prevent tissue section detachment during the retrieval process. | Essential due to the harsh nature of enzymatic treatment [30] [47]. |
Detailed PIER Protocols
PIER Using Trypsin
Protocol Steps:
- Deparaffinization and Rehydration: Follow standard IHC protocols to deparaffinize FFPE tissue sections in xylene and rehydrate through a graded ethanol series (e.g., 100%, 95%, 70%) to distilled water [6].
- Trypsin Solution Preparation: Prepare a 0.05% to 0.1% trypsin solution in a buffer, such as PBS or Tris-HCl. Critically, adjust the pH of the solution to 7.6 using NaOH [45]. The solution should be pre-warmed to 37°C before use.
- Enzymatic Digestion: Pipette the pre-warmed trypsin solution onto the tissue sections, ensuring complete coverage.
- Incubation: Incubate the slides in a humidity chamber at 37°C for 10 to 40 minutes [45]. Note: The optimal incubation time must be determined empirically and can be extended for certain worn-out tissues.
- Termination: Rinse the slides thoroughly with PBS or TBS to stop the enzymatic digestion.
- Continuation: Proceed with the standard IHC staining protocol, starting with blocking and primary antibody incubation.
PIER Using Proteinase K
Protocol Steps:
- Deparaffinization and Rehydration: As described in Section 5.1, Step 1.
- Proteinase K Solution Preparation: Prepare a working solution of Proteinase K at a concentration of 20 µg/mL in the recommended buffer (e.g., Tris-HCl with CaCl₂) or in PBS/TBS [45] [15]. An alternative protocol suggests a higher concentration of 0.6 units/mL [15].
- Enzymatic Digestion: Apply the Proteinase K working solution to cover the tissue sections.
- Incubation: Incubate the slides in a humidity chamber at 37°C for 20 minutes [45]. Other protocols use 15 minutes at 37°C [15].
- Termination and Cooling: Tap off the enzyme solution and rinse the slides thoroughly with TBS or PBS. A cooling-off period of 10 minutes at room temperature may follow [15].
- Continuation: Continue with the standard IHC staining protocol.
Optimization and Troubleshooting
Optimizing PIER is mandatory for achieving specific staining while preserving tissue integrity. The most critical variables are the enzyme concentration and incubation time. Over-digestion will damage morphology, while under-digestion will yield weak or false-negative signals [28]. It is strongly recommended to perform an initial optimization matrix using a range of times (e.g., 5, 10, 20, 30 minutes) with a fixed, standard concentration [28]. The fixation method and duration also significantly impact the required digestion time; longer fixation typically requires longer enzymatic treatment [30]. If poor staining persists with one enzyme, testing an alternative enzyme (e.g., switching from Trypsin to Proteinase K or pepsin) is advised [7]. Finally, the use of appropriate positive and negative controls is essential to confirm that the observed staining is specific and to rule out artifacts introduced by the retrieval process [46] [28].
From Problem to Solution: Troubleshooting Staining Artifacts and Optimizing Your Protocol
Under-retrieval is a primary cause of weak or absent signal in immunohistochemistry (IHC), occurring when the antigen retrieval step fails to adequately unmask epitopes obscured by formalin fixation. This technical challenge can lead to false-negative results, compromising experimental validity and reproducibility. Within the broader research on optimizing Heat-Induced Epitope Retrieval (HIER) versus Proteolytic-Induced Epitope Retrieval (PIER), effectively diagnosing and addressing under-retrieval is fundamental for researchers, scientists, and drug development professionals who rely on accurate protein localization and quantification. This guide provides a structured approach to identify and correct under-retrieval, ensuring maximal signal intensity and data quality.
Understanding the Root Causes of Under-Retrieval
Formalin fixation creates methylene bridges that cross-link proteins, masking epitopes and preventing antibody binding [48] [6]. Antigen retrieval methods, including HIER and PIER, reverse this masking. Under-retrieval occurs when the retrieval intensity is insufficient to break a critical number of these cross-links.
The choice between HIER and PIER is not universal; some epitopes require the specific action of one method. For instance, a 2024 study on osteoarthritic cartilage found that for detecting the cartilage intermediate layer protein 2 (CILP-2), PIER using proteinase K and hyaluronidase produced superior results compared to HIER or a combination of both methods [17] [24]. In this case, the application of heat in HIER not only reduced positive staining but also frequently caused section detachment [17]. This demonstrates that under-retrieval for a specific target can be method-dependent.
The diagram below outlines a systematic workflow for diagnosing the cause of weak or absent IHC signal.
Quantitative Comparison of Retrieval Methods
The following tables consolidate key experimental data from published studies to guide the initial selection and optimization of antigen retrieval methods.
Table 1: Comparative performance of antigen retrieval methods in different tissue types. Adapted from [17] [14].
| Tissue Type | Target Antigen | Optimal Method | Key Findings |
|---|---|---|---|
| Osteoarthritic Cartilage | CILP-2 | PIER (Proteinase K + Hyaluronidase) | PIER yielded the most abundant staining; HIER caused frequent section detachment [17]. |
| Mouse Decalcified Joint | p65, IL-1β, IL-6, IRF5 | Trypsin (PIER) or Improved Water Bath (IWB) | Pressure cooking (HIER) resulted in severe tissue detachment; Trypsin maintained better morphology [14]. |
Table 2: Optimization parameters for Heat-Induced Epitope Retrieval (HIER). Data from [49] [6].
| Parameter | Options | Optimization Guidelines |
|---|---|---|
| Buffer pH | Acidic (pH 6.0), Neutral (pH 7.0-7.6), Basic (pH 8.0-9.0) | Begin testing with citrate (pH 6.0) and Tris-EDTA (pH 9.0) [49] [6]. |
| Incubation Time | 1-20 minutes (pressure cooker), 20 minutes (microwave/steamer) | Use a time matrix (e.g., 1, 5, 15 min) to find the optimal duration [49]. |
| Temperature | 95°C - 100°C (microwave, steamer), ~120°C (pressure cooker) | Higher temperatures (pressure cooking) are more efficient but risk tissue damage [14] [6]. |
Table 3: Enzymatic conditions for Proteolytic-Induced Epitope Retrieval (PIER). Data from [17] [50].
| Enzyme | Working Concentration | Typical Digestion Conditions | Notes |
|---|---|---|---|
| Trypsin | 0.05% to 0.1% | 37°C for 10-40 minutes [50] | pH should be adjusted to 7.6-8.0 [50]. |
| Proteinase K | 20-30 µg/mL | 37°C for 20-90 minutes [17] [50] | Used in combination with hyaluronidase for cartilage matrix [17]. |
| Pepsin | 0.4% | 37°C for 30-180 minutes [50] | Suitable for more difficult epitopes. |
Experimental Protocols for Diagnosing and Correcting Under-Retrieval
Systematic Protocol for HIER Optimization
This protocol provides a stepwise method to optimize HIER, which is critical as its success is highly dependent on buffer pH, time, and temperature [49].
- Slide Preparation: Deparaffinize and rehydrate tissue sections using standard xylene and ethanol series [17].
- Buffer Selection: Prepare a minimum of three different retrieval buffers: 10 mM Sodium Citrate (pH 6.0), 1 mM EDTA (pH 8.0), and 10 mM Tris-EDTA (pH 9.0) [6].
- Heat Application: Using a preheated pressure cooker, microwave, or steamer, incubate slides in the retrieval buffer. For initial testing, a time matrix is crucial.
- Cooling: After heat treatment, remove the container from the heat source and cool it by running cold tap water over it for 10 minutes. This allows the reformed antigenic sites to stabilize [6].
- Immunostaining: Proceed with standard IHC steps: peroxidase blocking, blocking with serum or protein, primary antibody incubation, secondary antibody application, and chromogenic detection [17] [51].
- Validation: Always include a no-retrieval control and a known positive tissue control to contextualize the results and confirm that HIER is necessary [49] [52].
Systematic Protocol for PIER Optimization
PIER is a harsher method that can damage tissue morphology if over-optimized, but it is essential for certain epitopes and tissues like cartilage [17] [50].
- Slide Preparation: Deparaffinize and rehydrate tissue sections as above.
- Enzyme Solution Preparation:
- Enzymatic Digestion: Pipette the enzyme solution onto the slides and incubate in a humidified chamber at 37°C.
- Reaction Termination: Immerse the slides in distilled water to stop the enzymatic reaction.
- Immunostaining: Continue with the standard IHC protocol as described in the HIER section.
The Scientist's Toolkit: Key Research Reagent Solutions
Table 4: Essential reagents for antigen retrieval optimization and their functions.
| Reagent / Kit | Function | Specific Examples |
|---|---|---|
| Citrate Buffer (pH 6.0) | A common acidic buffer for HIER; ideal for many nuclear and cytoplasmic antigens [6]. | Citrate Buffer Powder (Bosterbio, AR0024) [50]. |
| Tris-EDTA Buffer (pH 9.0) | A common basic buffer for HIER; often superior for membrane proteins and some transcription factors [6]. | Abcam Tris-EDTA Buffer Kit [6]. |
| Proteinase K | A broad-spectrum serine protease used in PIER to digest cross-linking proteins [17]. | 20-30 µg/mL in Tris/HCl for cartilage tissue [17]. |
| Trypsin | A protease that cleaves peptide bonds; used for a wide range of epitopes in PIER [50]. | 0.05-0.1% solution, pH adjusted to 7.6-8.0 [50]. |
| Universal HIER Kit | A pre-formulated retrieval solution designed to work with a wide array of antibodies, reducing optimization time [6]. | Abcam Universal Heat-mediated Antigen Retrieval Reagent Kit [6]. |
| Polymer-Based Detection System | Secondary antibody systems conjugated to polymer-HRP, offering high sensitivity and reduced non-specific background [51]. | Dako REAL EnVision Detection System [17] [51]. |
Flowchart for Method Selection and Advanced Troubleshooting
When standard optimizations fail, a more strategic approach is required. The following diagram guides the selection and advanced troubleshooting of HIER and PIER methods, incorporating key decision points based on tissue type and antigen characteristics.
Antigen retrieval is a critical, yet often problematic, step in immunohistochemistry (IHC) that researchers must master to obtain reliable data. While essential for restoring antibody access to epitopes masked by formalin fixation, the process itself can introduce significant artifacts and elevated background staining if not properly controlled [53] [28]. The fundamental challenge lies in the fact that the same mechanisms that unmask target epitopes—whether through heat-induced breakdown of protein crosslinks or enzymatic digestion of surrounding proteins—can also expose non-target epitopes and disrupt tissue morphology, leading to compromised results [54] [55]. For researchers and drug development professionals working with precious clinical samples or evaluating therapeutic targets, understanding how to troubleshoot and resolve these issues is paramount to generating quantitatively accurate and reproducible IHC data, particularly within the broader context of optimizing Heat-Induced Epitope Retrieval (HIER) versus Proteolytic-Induced Epitope Retrieval (PIER) methodologies.
The transition from suboptimal to high-quality staining hinges on recognizing the visual hallmarks of over-retrieval and implementing systematic corrections. The following workflow outlines the critical decision points for diagnosing and resolving high background staining.
Understanding the Root Causes: Mechanisms of Over-Retrieval
How Over-Retrieval Creates Background Staining
The phenomenon of high background staining following antigen retrieval stems from two primary mechanisms: excessive unmasking of non-target epitopes and physical degradation of tissue integrity. During Heat-Induced Epitope Retrieval (HIER, excessive heat or prolonged heating can denature proteins beyond the optimal point, causing unfolding that exposes hydrophobic regions normally buried within the protein's native structure [53] [28]. These exposed hydrophobic areas promote non-specific antibody binding through hydrophobic interactions, creating diffuse cytoplasmic staining that obscures specific signal localization. Furthermore, overheating can damage tissue morphology, making cellular compartments less distinct and facilitating antibody trapping in structurally compromised areas [54].
In Proteolytic-Induced Epitope Retrieval (PIER), the primary risk stems from over-digestion of tissue proteins. When proteolytic enzymes like proteinase K, trypsin, or pepsin are applied at excessive concentrations or for prolonged durations, they can create artificial epitopes through partial protein digestion or expose charged residues that promote ionic interactions with antibody molecules [54] [55]. This excessive proteolysis may also create holes in the tissue architecture where antibodies become physically trapped, generating punctate staining patterns that lack biological relevance [8]. The visual manifestations of these processes provide critical diagnostic clues for identifying the specific type of over-retrieval at play, as detailed in the table below.
Table 1: Diagnostic Features of Over-Retrieval Artifacts
| Retrieval Type | Visual Cues | Tissue Morphology Impact | Common Antigens Affected |
|---|---|---|---|
| HIER Over-Retrieval | Diffuse cytoplasmic staining, elevated background across all tissue compartments [28] | Tissue detachment from slides, nuclear puffiness, loss of subcellular detail [54] | Nuclear antigens (p27, Ki-67), membrane proteins [53] [54] |
| PIER Over-Retrieval | Punctate staining patterns, irregular staining at tissue edges [55] | Holes in tissue section, loss of glandular architecture, friable tissue [8] | Extracellular matrix proteins (collagen, fibronectin) [54] |
The Delicate Balance: Epitope Retrieval and Background Formation
The relationship between optimal signal retrieval and background generation follows a predictable trajectory that researchers must recognize. Initially, as retrieval intensity increases from insufficient to optimal conditions, target-specific signal improves while background remains minimal. However, once the retrieval intensity surpasses the optimal point, background staining increases rapidly while specific signal quality deteriorates [53] [28]. This creates a narrow window where both specific signal and low background can be achieved—the precise target of method optimization.
The following diagram illustrates the relationship between retrieval intensity and staining quality, highlighting the narrow optimization window.
Systematic Troubleshooting Approaches
Troubleshooting HIER-Induced Background
When faced with high background staining potentially caused by HIER over-retrieval, a systematic approach to adjusting key parameters is essential:
Reduce Heat Exposure: The duration and intensity of heat application are primary levers for control. For microwave-based methods, reduce heating time from 20 minutes to 10-15 minutes, or implement intermittent heating cycles (e.g., 4 minutes at 95°C followed by cooling, then repeated) [54] [6]. When using a pressure cooker, decrease the time at full pressure from 3 minutes to 1-2 minutes [28]. Consider switching from a pressure cooker (120°C) to a water bath (92-95°C) or steamer (95-100°C) for gentler heat application [6] [28].
Optimize Retrieval Buffer pH: The pH of the retrieval buffer significantly influences which epitopes are unmasked and which charged residues become exposed. Test a range of pH conditions using citrate buffer (pH 6.0), Tris-EDTA (pH 9.0), or EDTA (pH 8.0) [54] [6]. For many nuclear antigens like Ki-67 and ER, higher pH buffers (pH 8-9) provide superior signal-to-noise ratio, while some membrane antigens may respond better to neutral pH conditions [53] [54].
Include Appropriate Controls: Always run a no-primary antibody control to detect secondary antibody non-specific binding, and a no-retrieval control to establish the baseline improvement from retrieval [28]. When optimizing, use a matrix approach that tests multiple time and pH combinations on serial sections of the same tissue block to directly compare outcomes [28].
Addressing PIER-Related Over-Digestion
PIER over-retrieval requires different corrective strategies focused on enzymatic activity control:
Titrate Enzyme Concentration and Time: Reduce proteinase K concentration from 20μg/mL to 5-10μg/mL, or decrease trypsin concentration from 0.1% to 0.05% [55]. Shorten digestion time from 30 minutes to 10-15 minutes at 37°C [54] [6]. Always prepare fresh enzyme solutions for consistency.
Evaluate Alternative Enzymes: If trypsin or proteinase K causes excessive damage, test gentler enzymes like pepsin (0.4% for 30-180 minutes at 37°C) for extracellular matrix antigens or bromelain for interstitial antigens like fibronectin and collagen [54] [55].
Precisely Control Digestion Conditions: Maintain strict temperature control at 37°C throughout digestion using a calibrated incubator rather than bench-top application [54]. Terminate reactions promptly by transferring slides to tap water for thorough washing [54].
Quantitative Optimization Framework
Systematic optimization using a structured experimental approach is the most reliable path to resolving background issues. The following matrix provides a quantitative framework for simultaneously testing multiple retrieval parameters, enabling researchers to identify the optimal conditions that maximize signal while minimizing background.
Table 2: Antigen Retrieval Optimization Matrix for HIER
| Time | Citrate Buffer (pH 6.0) | EDTA Buffer (pH 8.0) | Tris-EDTA (pH 9.0) |
|---|---|---|---|
| 5 minutes | Specific Signal: ++\nBackground: +\nMorphology: +++ | Specific Signal: ++\nBackground: +\nMorphology: +++ | Specific Signal: +++\nBackground: +\nMorphology: +++ |
| 10 minutes | Specific Signal: +++\nBackground: ++\nMorphology: ++ | Specific Signal: ++++\nBackground: ++\nMorphology: ++ | Specific Signal: ++++\nBackground: ++\nMorphology: ++ |
| 15 minutes | Specific Signal: +++\nBackground: +++\nMorphology: + | Specific Signal: ++++\nBackground: +++\nMorphology: + | Specific Signal: +++\nBackground: ++++\nMorphology: + |
Scoring: + to ++++ (lowest to highest). Example data based on typical optimization patterns for nuclear antigens [54] [28].
Detailed Experimental Protocols for Background Reduction
Optimized HIER Protocol for Sensitive Tissues
This protocol is specifically designed to minimize background while maintaining strong specific signal in delicate tissues prone to over-retrieval:
Deparaffinize and rehydrate tissue sections using standard xylene and graded ethanol series [6].
Prepare retrieval buffer - Tris-EDTA (10 mM Tris base, 1 mM EDTA, 0.05% Tween 20, pH 9.0) provides excellent results for most nuclear antigens with minimal background [6].
Heat using a water bath - Place slides in preheated buffer at 92-95°C for 10-12 minutes. The water bath provides more uniform heating than microwaves, reducing localized over-heating [28].
Cool gradually - After retrieval, allow slides to remain in the buffer as it cools to room temperature for 15-25 minutes. This gradual cooling promotes proper epitope re-folding [6].
Proceed with staining - Wash slides in PBS before continuing with peroxidase blocking and antibody application [6].
Gentle PIER Protocol for Fragile Antigens
For antigens that require enzymatic retrieval but are prone to over-digestion artifacts:
Prepare working enzyme solution - Dilute proteinase K to 5-10μg/mL in 50 mM Tris/HCl, 5 mM CaCl₂ solution (pH 6.0) [8]. Keep on ice until use.
Apply to tissue sections - Pipette minimal volume needed to cover tissue and place in humidity chamber.
Incubate precisely - Transfer to 37°C incubator for exactly 8-12 minutes. Use a timer for consistency.
Stop reaction immediately - Transfer slides directly to Tris-buffered saline and rinse for 3×2 minutes to completely remove enzyme [54] [8].
Consider combinatorial approach - For challenging antigens, test sequential HIER (mild, 5 minutes at 95°C) followed by PIER (reduced concentration, 5 minutes) [8].
The Scientist's Toolkit: Essential Reagents for Troubleshooting
Table 3: Key Research Reagent Solutions for Background Troubleshooting
| Reagent/Catalog | Primary Function | Optimization Guidance |
|---|---|---|
| Citrate Buffer (pH 6.0) [6] | HIER buffer for antigens requiring acidic pH | First-line for many antigens; test if high background occurs with high-pH buffers |
| Tris-EDTA (pH 9.0) [6] | HIER buffer for nuclear antigens | Superior for ER, Ki-67; often provides cleaner background than citrate |
| EDTA (pH 8.0) [54] | HIER buffer for challenging nuclear targets | More effective than citrate for many nuclear antigens; may require morphology trade-offs |
| Proteinase K [8] [55] | Proteolytic enzyme for PIER | Use low concentrations (5-10μg/mL); monitor tissue morphology closely |
| Trypsin [54] [55] | Proteolytic enzyme for PIER | Effective at 0.05-0.1%; requires precise pH adjustment to 7.6 |
| Universal AR Reagents [28] | Pre-optimized for multiple targets | Reduce optimization time; particularly valuable for screening antibodies |
Resolving high background staining in IHC requires recognizing that antigen retrieval exists on a continuum where both under-retrieval and over-retrieval produce suboptimal results. The most successful approaches implement systematic optimization of time, temperature, pH, and buffer composition while recognizing that the optimal balance point is unique to each antigen-antibody pair and tissue type [28]. By applying the detailed protocols and troubleshooting frameworks presented here, researchers can transform problematic IHC staining into quantitatively reliable data, advancing both basic research and drug development objectives through more reproducible and interpretable immunohistochemical analysis.
Antigen retrieval is a critical preparatory step in immunohistochemistry (IHC) that reverses the epitope masking caused by formalin fixation [28]. While essential for effective antibody binding, the process itself presents a significant challenge: preserving optimal tissue morphology. The very methods that expose antigens—heat-induced epitope retrieval (HIER) and proteolytic-induced epitope retrieval (PIER)—can damage tissue integrity, leading to artifactual staining, section detachment, and compromised interpretation [17] [56]. This application note details practical strategies for mitigating morphology loss during antigen retrieval, providing a framework for researchers to balance robust immunoreactivity with superior tissue preservation.
Mechanisms and Trade-offs of Antigen Retrieval
Formaldehyde fixation creates methylene bridges that cross-link proteins, thereby masking epitopes and limiting antibody access [28] [57]. Antigen retrieval aims to break these cross-links, but the two primary methods achieve this through different mechanisms, each with distinct implications for tissue integrity.
Heat-Induced Epitope Retrieval (HIER) uses heated buffer solutions (e.g., citrate pH 6.0, Tris-EDTA pH 9.0) to break protein cross-links, believed to reverse some cross-links and restore the epitope's secondary or tertiary structure [28] [30]. While generally gentler on morphology than enzymatic methods, its main risks include tissue section detachment from slides due to vigorous boiling and potential destruction of heat-labile epitopes, particularly in poorly glycosylated proteins [17] [57].
Proteolytic-Induced Epitope Retrieval (PIER) employs enzymes like proteinase K, trypsin, and pepsin to digest proteins and cleave cross-linking peptides that may be masking the epitope [28] [30]. This method is harsher on tissue morphology; over-digestion can destroy cellular ultrastructure, damage the antigen of interest, and lead to excessive hydrolysis [28] [56].
Table 1: Primary Risks of Antigen Retrieval Methods to Tissue Morphology
| Method | Primary Mechanism | Main Morphological Risks |
|---|---|---|
| HIER | Breakage of cross-links via heat and buffer [28] [30] | Tissue detachment from slides [17]; potential damage to heat-labile antigens [17] |
| PIER | Enzymatic digestion of cross-linking peptides [28] [30] | Over-digestion leading to loss of cellular detail and antigen destruction [28] [56] |
Strategic Optimization to Minimize Tissue Damage
General Foundational Practices
- Adhesive Slides: Use poly-L-lysine or other positively-charged adhesive slides to prevent tissue detachment during HIER's heating and cooling cycles [30].
- Fixation Control: Standardize fixation type and duration, as over-fixation increases cross-linking and necessitates harsher retrieval, raising damage risk [30].
- Systematic Optimization: Optimize retrieval conditions using a matrix approach, testing different combinations of time, temperature, and pH for each new antibody-antigen pair [28] [57].
HIER-Specific Mitigation Strategies
- Precise Temperature Control: Avoid uncontrolled boiling by using purpose-built scientific microwaves, water baths, or pressure cookers that provide consistent, uniform heating to minimize tissue damage [30].
- Buffer and pH Optimization: Test various buffer pH levels, as the optimal pH is often antigen-dependent and can significantly influence staining quality and morphology preservation [28] [57] [30].
- Controlled Cooling: Allow slides to cool in the retrieval buffer after heating to avoid thermal shock that can damage tissue morphology [28].
PIER-Specific Mitigation Strategies
- Enzyme Titration and Timing: Precisely optimize enzyme concentration and incubation time, as the optimal conditions depend on the specific enzyme, tissue type, and fixation length [56] [30].
- Temperature Control: Perform enzymatic digestions at a consistent 37°C to ensure predictable enzyme activity [56].
- Reaction Termination: Immediately terminate the enzymatic reaction by transferring slides to cold water or buffer and rinsing thoroughly after the incubation period to prevent over-digestion [57].
Table 2: Troubleshooting Guide for Morphology Preservation
| Problem | Potential Causes | Corrective Actions |
|---|---|---|
| Tissue Detachment | Insufficient slide adhesion; excessive boiling during HIER [17] | Use charged/coated slides; ensure slides are fully dried before retrieval; reduce heating vigor [30]. |
| Over-digestion (PIER) | Enzyme concentration too high; incubation time too long [28] | Titrate enzyme concentration; reduce incubation time in 5-minute increments [56]. |
| Excessive Tissue Damage (HIER) | Temperature too high; prolonged heating [17] | Lower temperature (e.g., 95°C vs. pressure cooker 120°C); reduce heating duration [28]. |
| Poor Staining & Poor Morphology | Unsuitable retrieval method; suboptimal buffer pH [57] | Switch from HIER to PIER or vice versa; test a pH gradient (e.g., pH 6.0, 8.0, 9.0) [7]. |
Experimental Protocols for Comparative Analysis
Case Study: Optimizing Retrieval for a Cartilage Glycoprotein
A 2024 study on osteoarthritic cartilage highlights the need for protocol adaptation to preserve morphology and enhance staining [17] [8]. The dense extracellular matrix and low abundance of the target protein (CILP-2) required careful optimization.
- Sample Preparation: Human OA cartilage samples were fixed in 10% buffered formalin (<3 weeks), decalcified, and embedded in paraffin. Sections were cut at 4 µm and mounted on adhesion slides [17] [8].
- Protocol Comparison: Four retrieval conditions were tested on sequential sections for direct comparison [17] [8]:
- HIER Only: Heated in decloaking solution at 95°C for 10 minutes [17].
- PIER Only: Incubated with 30 µg/mL Proteinase K in Tris/HCl (pH 6.0) for 90 minutes at 37°C, followed by 0.4% hyaluronidase for 3 hours at 37°C [17].
- Combined HIER/PIER: Sequential treatment with the above HIER and PIER protocols [17].
- Control: No antigen retrieval [17].
- Key Findings and Morphological Outcome: The PIER-only protocol yielded the best CILP-2 staining. The combined HIER/PIER approach did not improve results and caused frequent section detachment, demonstrating the morphological risk of overly harsh combined treatments [17]. This underscores that a simpler, well-optimized single method is often superior for preserving tissue integrity.
Decision Workflow for Method Selection
The following workflow provides a logical pathway for selecting and optimizing an antigen retrieval method to minimize tissue damage.
The Scientist's Toolkit: Essential Reagents and Materials
Table 3: Key Research Reagent Solutions for Antigen Retrieval
| Item | Function/Application | Specific Examples |
|---|---|---|
| Adhesive Slides | Prevents tissue detachment during HIER [30] | Poly-L-lysine, APES, or positively-charged slides [30] |
| HIER Buffers | Solution for heat-based unmasking; pH critical for success [57] [30] | Sodium Citrate (pH 6.0), Tris-EDTA (pH 9.0), EDTA (pH 8.0) [7] [57] |
| Proteolytic Enzymes | Enzymes for PIER; choice depends on antigen location [57] | Trypsin (0.05-0.1%), Proteinase K (e.g., 20 µg/mL), Pepsin (0.4%) [56] |
| Heating Devices | Appliances for HIER; must provide consistent, controlled heat [28] [30] | Scientific microwave, pressure cooker, vegetable steamer, water bath [28] |
| Neutral Buffers | For diluting enzymes and washing steps [7] | Phosphate-Buffered Saline (PBS), Tris-Buffered Saline [7] |
Preventing tissue damage during antigen retrieval is an achievable goal that hinges on systematic optimization and a deep understanding of the trade-offs involved. There is no universal solution; the optimal protocol must be empirically determined for each specific antigen, tissue, and fixation combination. By employing a strategic approach—starting with gentler HIER, meticulously optimizing conditions using a matrix, and using PIER for more stubborn epitopes—researchers can successfully navigate the balance between powerful antigen detection and the impeccable preservation of tissue morphology that is essential for accurate biological interpretation.
Antigen retrieval is a critical pre-analytical step in immunohistochemistry (IHC) that reverses the epitope masking caused by formalin fixation [28] [30] [2]. Although fixation is essential for preserving tissue morphology, the process of cross-linking proteins can obscure antigenic sites, rendering them inaccessible to primary antibodies [28]. The development of robust antigen retrieval methods has significantly improved the sensitivity and reproducibility of immunostaining in formalin-fixed, paraffin-embedded (FFPE) tissues, making it one of the most important factors affecting IHC results [30].
This application note establishes a systematic framework for optimizing antigen retrieval parameters, focusing specifically on the strategic testing of time, temperature, and pH matrices. Within the broader context of Heat-Induced Epitope Retrieval (HIER) versus Proteolytic-Induced Epitope Retrieval (PIER) research, this protocol provides researchers and drug development professionals with standardized methodologies to enhance antibody binding efficiency while preserving tissue integrity [58] [59]. The strategic optimization outlined here is particularly valuable for novel antibody validation, diagnostic assay development, and investigative pathology where reproducible immunostaining is paramount.
Theoretical Background: HIER vs. PIER Mechanisms
Fundamental Principles of Antigen Retrieval
Formalin fixation creates methylene bridges between amino acid residues, leading to cross-linking that physically blocks antibody access to epitopes [28] [2]. Antigen retrieval methods aim to break these cross-links without damaging tissue morphology or the antigenic sites themselves. The two primary approaches—HIER and PIER—operate through distinct mechanisms and are suited to different applications [59].
Heat-Induced Epitope Retrieval (HIER) utilizes elevated temperatures (typically 95-120°C) in specific buffer solutions to disrupt protein cross-links through thermal energy [28] [60]. The exact mechanism remains partially characterized but is believed to involve:
- Breaking of formalin-induced cross-links between epitopes and unrelated proteins [30]
- Extraction of diffusible blocking proteins [30]
- Calcium ion chelation, as many retrieval buffers contain EDTA [2]
- Rehydration of tissue sections allowing better antibody penetration [30]
- Restoration of the epitope's secondary or tertiary structure [28]
Proteolytic-Induced Epitope Retrieval (PIER) employs enzymatic digestion using proteases such as proteinase K, trypsin, or pepsin to cleave peptide bonds and remove obstructive proteins [28] [8]. This method typically operates at 37°C with incubation periods ranging from 10-120 minutes depending on the tissue and fixation duration [30] [2].
Comparative Analysis of HIER and PIER
The following dot graph illustrates the key procedural differences and decision points for selecting between HIER and PIER methods:
Materials and Reagent Solutions
Research Reagent Solutions
The following table details essential reagents and materials required for implementing the strategic optimization framework:
Table 1: Essential Research Reagents for Antigen Retrieval Optimization
| Reagent Category | Specific Examples | Function and Application Notes |
|---|---|---|
| HIER Buffers | 10 mM Sodium Citrate (pH 6.0) | Low-pH buffer effective for many nuclear antigens [6] [60] |
| Tris-EDTA (pH 8.0-9.0) | High-pH buffer often more effective for membrane proteins [30] [6] | |
| EDTA (pH 8.0) | Chelating buffer suitable for low abundance proteins [60] | |
| PIER Enzymes | Proteinase K | Broad-spectrum serine protease; concentration typically 5-30 µg/mL [8] |
| Trypsin | Serine endopeptidase; optimal at pH 7.8 [30] [2] | |
| Pepsin | Aspartic protease; functions well in acidic environment [30] | |
| Equipment | Pressure Cooker | Delivers 120°C for rapid retrieval (1-5 minutes) [28] [6] |
| Water Bath | Maintains 92-95°C for 5-10 minutes [28] [6] | |
| Microwave | Domestic or scientific; 20 minutes at 98°C [6] | |
| Steamer | Maintains 95-100°C for 20-40 minutes [6] [60] | |
| Control Reagents | Universal Retrieval Reagents | Pre-formulated solutions covering multiple pH ranges [28] |
| Antibody Diluents | pH-adjusted solutions to enhance antibody affinity [28] |
Experimental Design and Matrix Setup
Strategic Optimization Framework
A systematic approach to antigen retrieval optimization requires testing multiple parameters in a matrix format. This framework enables researchers to efficiently identify optimal conditions for specific antibody-antigen combinations while maintaining tissue morphology.
Table 2: Experimental Matrix for HIER Optimization (Time × pH)
| Time \ pH | Acidic (pH 5.0) | Neutral (pH 7.0) | Basic (pH 9.5) |
|---|---|---|---|
| 1 minute | Slide #1 | Slide #2 | Slide #3 |
| 5 minutes | Slide #4 | Slide #5 | Slide #6 |
| 10 minutes | Slide #7 | Slide #8 | Slide #9 |
Note: This matrix should be compared against a control slide with no HIER treatment [28]. Similar matrices can be developed for temperature optimization using ranges from 90-120°C depending on the heating method [28] [30].
Workflow for Comprehensive Antigen Retrieval Optimization
The following flowchart outlines the complete experimental workflow for systematic optimization of antigen retrieval conditions:
Detailed Experimental Protocols
HIER Protocol Using Pressure Cooker
The pressure cooker method provides rapid and uniform heating, making it highly effective for difficult-to-retrieve antigens [6].
Materials:
- Domestic stainless steel pressure cooker
- Hot plate
- Slide rack capable of withstanding high temperature
- Antigen retrieval buffer (citrate pH 6.0, Tris-EDTA pH 9.0, or EDTA pH 8.0)
Procedure:
- Add appropriate antigen retrieval buffer to the pressure cooker [6].
- Place the pressure cooker on a hot plate at full power without securing the lid [6].
- While waiting for the buffer to boil, deparaffinize and rehydrate tissue sections using standard xylene and ethanol series [6].
- Once boiling, transfer slides from tap water to the pressure cooker using forceps [6].
- Secure the pressure cooker lid according to manufacturer's instructions [6].
- Once full pressure is reached, time for 3 minutes [6].
- After 3 minutes, turn off the hotplate and place the pressure cooker in an empty sink.
- Activate the pressure release valve and run cold water over the cooker [6].
- Once depressurized, open the lid and run cold water into the cooker for 10 minutes to cool slides [6].
- Continue with immunohistochemical staining protocol.
HIER Protocol Using Microwave
Microwave-based retrieval offers precise temperature control in scientific models, though domestic microwaves may create hot and cold spots [6].
Materials:
- Scientific microwave (preferred) or domestic microwave (850W)
- Microwaveable vessel with slide rack
- Antigen retrieval buffer
Procedure:
- Deparaffinize and rehydrate tissue sections [6].
- Add sufficient antigen retrieval buffer to microwaveable vessel to cover slides by several centimeters [6].
- Place slides in the buffer-filled vessel [6].
- For domestic microwaves: Heat at full power until boiling, then boil for 20 minutes [6].
- For scientific microwaves: Program to maintain 98°C for 20 minutes after reaching temperature [6].
- Monitor buffer levels throughout the procedure and add distilled water if necessary to prevent drying [6].
- After 20 minutes, remove vessel and run cold tap water into it for 10 minutes to cool slides [6].
- Continue with immunohistochemical staining protocol.
PIER Protocol Using Proteinase K
Enzymatic retrieval is particularly useful for densely structured tissues or specific glycoprotein antigens that may not respond well to heat-induced methods [8].
Materials:
- Proteinase K (0.6 units/mL working solution) [15]
- Humidity chamber
- Tris/HCl buffer with CaCl₂ (pH 6.0) [8]
- Water bath or incubator maintaining 37°C
Procedure:
- Deparaffinize and rehydrate tissue sections through xylene and graded ethanol series to water.
- Pipette Proteinase K working solution onto slides, ensuring complete tissue coverage [15].
- Place slides in a humidity chamber and incubate for 15 minutes at 37°C [15].
- For challenging tissues or heavily cross-linked antigens, extend incubation time up to 90 minutes [8].
- Terminate digestion by transferring slides to Tris-buffered saline (TBS) at room temperature for 10 minutes [15].
- Rinse slides gently with distilled water before proceeding with immunohistochemical staining.
Combined HIER/PIER Approach
For exceptionally challenging antigens, a sequential combination of heat and enzymatic retrieval may be necessary, though this approach requires careful optimization to prevent tissue damage [8].
Procedure:
- Perform standard HIER protocol as described in sections 5.1 or 5.2.
- Cool slides to room temperature.
- Apply PIER protocol as described in section 5.3, potentially with reduced enzyme concentration or incubation time.
- Note: Combined methods may increase tissue detachment from slides; use charged or coated slides to improve adhesion [8].
Results Interpretation and Quality Control
Assessment Criteria for Optimization Outcomes
Successful antigen retrieval optimization requires systematic evaluation using multiple criteria to balance signal intensity with preservation of tissue morphology:
- Signal Intensity: Assess specific staining at the expected cellular localization (nuclear, cytoplasmic, membrane) [28].
- Background Staining: Evaluate non-specific background; excessive background may indicate over-retrieval [2].
- Tissue Morphology: Check for preservation of cellular structure; deterioration suggests excessive retrieval conditions [28] [8].
- Specificity: Verify that staining pattern matches expected antigen distribution [30].
Essential Experimental Controls
Appropriate controls are mandatory for validating antigen retrieval specificity and avoiding artifactual results:
- No Retrieval Control: Tissue section processed without antigen retrieval to establish baseline staining [28] [58].
- Negative Control: Section incubated with isotype control or without primary antibody to identify non-specific secondary antibody binding [15] [2].
- Positive Control: Tissue with known expression of the target antigen to confirm protocol effectiveness [58] [2].
- Specificity Controls: When available, use blocking peptides or knockout tissues to verify antibody specificity [30] [2].
Applications and Context-Specific Recommendations
Tissue-Specific Considerations
Different tissue types present unique challenges for antigen retrieval optimization:
- Cartilage Tissue: PIER methods may outperform HIER for certain matrix glycoproteins. A recent study on cartilage intermediate layer protein 2 (CILP-2) found proteinase K digestion more effective than heat-induced methods [8].
- Reproductive Tract Tissue: For murine female reproductive tract tissue, HIER at 80°C in citrate buffer provided optimal results for eosinophil protein and HSV-2 detection [15].
- Bone and Calcified Tissues: These may require extended retrieval times or specialized decalcification procedures prior to antigen retrieval [6].
Antigen-Specific Optimization Strategies
- Nuclear Antigens: Often respond well to citrate buffer at pH 6.0 with moderate heating times (10-15 minutes) [28].
- Membrane Proteins: Frequently require high-pH buffers (Tris-EDTA, pH 8.0-9.0) and longer retrieval times [30] [60].
- Phosphorylated Epitopes: May need specialized retrieval conditions to preserve post-translational modification recognition [61].
Troubleshooting Common Issues
- Weak or No Staining: Increase retrieval time, try higher pH buffer, or switch to pressure cooker method for higher temperature [58] [2].
- High Background: Reduce retrieval time, try lower pH buffer, or decrease enzyme concentration in PIER [2].
- Tissue Detachment: Use charged or coated slides, reduce retrieval temperature, or avoid boiling in microwave methods [6] [8].
- Over-retrieval Artifacts: Characterized by diffuse cytoplasmic staining; reduce time, temperature, or enzyme concentration [28].
The strategic optimization framework presented here provides a systematic methodology for evaluating antigen retrieval parameters through controlled testing of time, temperature, and pH matrices. By implementing this comprehensive approach, researchers can significantly enhance immunohistochemical staining reproducibility, particularly when developing assays for novel targets or validating antibodies for diagnostic applications.
Within the broader context of HIER versus PIER research, this protocol emphasizes an evidence-based approach to method selection, recognizing that while HIER generally offers higher success rates for most applications [28] [58], PIER remains valuable for specific antigens and tissue types [8]. The standardized methodologies and quality control measures outlined enable researchers to establish optimized antigen retrieval conditions that maximize antibody binding while preserving tissue morphology, ultimately supporting reliable and reproducible research and diagnostic outcomes.
In the context of optimizing antigen retrieval methods for immunohistochemistry (IHC), the implementation of proper controls is not merely a supplementary technique but a fundamental requirement for validating experimental outcomes. The critical choice between Heat-Induced Epitope Retrieval (HIER) and Proteolytic-Induced Epitope Retrieval (PIER) can significantly impact antibody binding efficiency and staining specificity, making robust control systems essential for accurate interpretation [62]. Antigen retrieval techniques aim to reverse the masking of epitopes caused by formalin fixation, which creates crosslinks that restrict antibody-target binding [41] [63]. Without appropriate controls, researchers cannot distinguish between true positive signals and artifacts introduced by the retrieval process itself, potentially compromising data reliability and experimental conclusions.
The complex matrix of tissues such as osteoarthritic cartilage further amplifies this need, as the dense extracellular environment inhibits antibody penetration and necessitates optimized retrieval protocols [17]. As the field advances toward automated IHC staining systems and standardized protocols, the role of controls becomes increasingly important for quality assurance and inter-laboratory reproducibility [64]. This application note provides detailed methodologies for implementing a comprehensive control system within the specific context of HIER versus PIER optimization research.
The Critical Function of Controls in IHC
Controls in IHC serve multiple essential functions: they verify the specificity of the antibody-antigen interaction, monitor technical procedural accuracy, and provide benchmarks for interpreting staining results. The implementation of appropriate controls is particularly crucial when comparing antigen retrieval methods because variables such as buffer pH, incubation time, and temperature can dramatically affect epitope availability and tissue morphology [65] [28].
Positive controls validate that all components of the IHC protocol are functioning correctly by using tissues or cells known to express the target antigen. These controls confirm protocol and reagent functionality and should ideally show consistent, expected staining patterns [62]. Negative controls eliminate the primary antibody or use an irrelevant antibody to assess background staining levels and identify non-specific binding [62]. Specificity controls provide additional verification that the observed staining results from the specific antibody-antigen interaction, which can be achieved through absorption tests or isotype controls.
The consequences of inadequate controls include false positive results due to non-specific staining or endogenous enzyme activity, and false negative results from insufficient antigen retrieval or improper antibody dilution [62]. These errors can lead to incorrect biological interpretations, especially when evaluating subtle differences in protein expression across experimental conditions. In the context of antigen retrieval optimization, controls must be implemented systematically to account for variations introduced by different HIER and PIER protocols.
Table 1: Types of Essential Controls in IHC Experiments
| Control Type | Purpose | Implementation Example | Interpretation |
|---|---|---|---|
| Positive Control | Validate protocol and reagent functionality | Tissue known to express target antigen | Expected staining confirms proper procedure |
| Negative Control | Assess background and non-specific staining | Omit primary antibody or use irrelevant antibody | No staining indicates specific signal |
| Specificity Control | Verify antibody binding specificity | Pre-absorb antibody with excess antigen | Significant reduction in staining confirms specificity |
| Retrieval Control | Evaluate antigen retrieval effectiveness | Include sample with no retrieval | Improved staining with retrieval demonstrates efficacy |
Quantitative Comparison of Control Performance
Recent studies have provided quantitative data on control performance in IHC applications. A 2025 study investigating a novel automated IHC staining system for ALK rearrangement detection in lung adenocarcinoma demonstrated excellent sensitivity (98.30%) and specificity (100%) when using appropriate controls [64]. This highlights how proper control implementation contributes to reliable diagnostic outcomes.
In a 2024 study comparing antigen retrieval methods for cartilage intermediate layer protein 2 (CILP-2) detection, researchers utilized a semi-quantitative staining assessment based on staining extent to evaluate four different protocols [17]. The study found that PIER using Proteinase K and hyaluronidase produced the best staining results for this particular glycoprotein, while HIER combined with PIER actually reduced staining quality and caused frequent section detachment [17] [24]. This finding underscores the antigen-specific nature of retrieval optimization and the importance of controls in making such determinations.
Table 2: Performance Comparison of Antigen Retrieval Methods for CILP-2 Detection
| Retrieval Method | Staining Extent | Tissue Preservation | Protocol Details | Advantages/Limitations |
|---|---|---|---|---|
| No Retrieval (Control) | Minimal | Excellent | N/A | Baseline for comparison; demonstrates need for retrieval |
| HIER | Moderate | Good | 95°C for 10 min in Decloaker solution | Standard approach; minimal tissue damage |
| PIER | Maximal | Moderate | 30 µg/mL Proteinase K (90 min) + 0.4% hyaluronidase (3 h) at 37°C | Optimal for CILP-2; longer protocol |
| HIER/PIER Combined | Reduced | Poor (frequent detachment) | Sequential application of both methods | Counterproductive effect; damages tissue integrity |
Statistical analysis of IHC results should incorporate measures of interobserver variability, especially when using semi-quantitative scoring systems. The 2025 ALK study utilized Cohen's Kappa statistics to compare diagnostic variability among five pathologists, providing a quantitative measure of interpretation consistency [64]. Such methodological rigor strengthens the validity of conclusions drawn from controlled IHC experiments.
Experimental Protocols for Control Implementation
Protocol for Controlled Comparison of HIER vs. PIER
Materials Required:
- Formalin-fixed, paraffin-embedded tissue sections
- Positive control tissue known to express target antigen
- Antigen retrieval buffers (acidic, neutral, basic)
- Proteinase K, trypsin, or pepsin for PIER
- Water bath, steamer, or microwave for HIER
- Primary antibody specific to target antigen
- Detection system (e.g., DAB chromogen)
- Hematoxylin counterstain
Procedure:
- Section Preparation: Cut 4μm thick sections from FFPE tissue blocks and mount on adhesive slides. Include both test tissues and positive control tissues on each slide batch.
Deparaffinization and Rehydration:
- Incubate slides in xylene (3 changes, 5 minutes each)
- Hydrate through graded ethanol series (100%, 95%, 70% - 2 minutes each)
- Rinse in distilled water
Antigen Retrieval Matrix Setup:
- Divide slides into groups for different retrieval conditions:
- Group 1: No retrieval (negative control for retrieval efficacy)
- Group 2: HIER with citrate buffer (pH 6.0), 95-100°C for 20-40 minutes [63]
- Group 3: HIER with Tris-EDTA buffer (pH 9.0), 95-100°C for 20-40 minutes [63]
- Group 4: PIER with Proteinase K (20μg/mL in TE buffer), 37°C for 10-20 minutes [41]
- Group 5: PIER with trypsin (0.05% working solution), 37°C for 10-20 minutes [41]
- After retrieval, cool slides for 20 minutes at room temperature
- Divide slides into groups for different retrieval conditions:
Immunostaining:
- Block endogenous peroxidase with 0.6% H₂O₂ for 15 minutes
- Wash in PBS (3 × 5 minutes)
- Block non-specific binding with protein block (5-10 minutes)
- Apply primary antibody at optimized dilution (overnight at 4°C)
- Include negative control slides without primary antibody
- Apply secondary antibody and detection system according to manufacturer's instructions
- Counterstain with hematoxylin, dehydrate, clear, and mount
Analysis and Interpretation:
- Compare staining intensity, distribution, and background across retrieval conditions
- Use positive controls to verify protocol efficacy
- Use negative controls to identify non-specific staining
- Employ semi-quantitative scoring (0-3+) for systematic comparison
Protocol for Automated IHC Quality Control
For laboratories utilizing automated IHC systems, the 2025 study on ALK testing provides a protocol for implementing controls in liquid form (CLFs) [64]:
Materials:
- Automated IHC staining system with quality control module (e.g., LYNX480 PLUS)
- CLFs from genetically modified cell lines (positive and negative)
- Primary antibody of interest
- Appropriate detection reagents
Procedure:
- Program the QC module to automatically apply CLFs onto slides before IHC staining
- Ensure proper drying and fixation of CLFs using the system's heater
- Proceed with automated deparaffinization, rehydration, and antigen retrieval
- Perform immunostaining according to established protocols
- Validate staining patterns in CLFs to confirm proper reagent performance
- Document all QC results for tracking and troubleshooting
Visualization of Workflows and Relationships
Experimental Control Workflow
The Scientist's Toolkit: Essential Research Reagents
Table 3: Essential Research Reagents for Controlled Antigen Retrieval Studies
| Reagent Category | Specific Examples | Function | Application Notes |
|---|---|---|---|
| Proteolytic Enzymes | Proteinase K, Trypsin, Pepsin | Digest cross-linking proteins to unmask epitopes in PIER | Concentration and incubation time must be optimized; may damage tissue morphology [41] [19] |
| HIER Buffers | Citrate (pH 6.0), Tris-EDTA (pH 9.0), Sodium Citrate | Restore epitope structure through heat-mediated reversal of cross-links | pH significantly affects retrieval efficiency; must be optimized for each antigen [65] [63] |
| Control Materials | Cell pellets, Tissue microarrays, Controls in Liquid Form (CLFs) | Verify staining protocol performance and antibody specificity | CLFs enable automated quality control without consuming scarce tissue [64] |
| Detection Systems | DAB chromogen, Hematoxylin counterstain | Visualize antibody binding and provide morphological context | Proper dilution and incubation time prevent background staining [62] |
| Validation Antibodies | Anti-CILP-2, Anti-ALK, Anti-p27 | Demonstrate antigen retrieval efficacy for specific targets | Supplier-recommended protocols provide starting points for optimization [17] [64] [28] |
The implementation of comprehensive control systems is fundamental to rigorous antigen retrieval optimization research. As demonstrated in studies comparing HIER and PIER methods, the optimal retrieval approach is highly dependent on the specific antigen-target pair and tissue type [17] [24]. By systematically employing positive, negative, and specificity controls according to the protocols detailed in this application note, researchers can confidently validate their findings and contribute to the advancement of reproducible IHC methodologies. The integration of automated quality control systems and standardized control materials represents the future of reliable immunohistochemical analysis in both research and diagnostic contexts [64].
HIER vs. PIER: An Evidence-Based Comparison for Strategic Method Selection
Within modern immunohistochemistry (IHC) laboratories, the analysis of formalin-fixed, paraffin-embedded (FFPE) tissues is fundamental to both diagnostic pathology and research. Formaldehyde fixation, while excellent for preserving tissue morphology, creates methylene cross-links between proteins that mask antigenic epitopes, thereby hindering antibody binding and detection [18] [32] [66]. To overcome this limitation, antigen retrieval techniques are a critical pre-analytical step. The two principal methodologies are Heat-Induced Epitope Retrieval (HIER) and Proteolytic-Induced Epitope Retrieval (PIER). The choice between HIER and PIER significantly impacts staining intensity, specificity, and tissue morphology [8] [67] [66]. This application note provides a direct comparison of these core techniques, framed within the broader thesis of optimizing IHC workflows for robust and reproducible results in research and drug development.
Mechanism of Action
The two methods employ fundamentally different mechanisms to unmask epitopes.
HIER: This method utilizes thermal energy, typically between 95°C and 120°C, in specific buffered solutions. The exact mechanism remains under investigation but is believed to involve the hydrolytic cleavage of the formaldehyde-induced cross-links, the unfolding of epitopes to restore their original conformation, and/or the chelation of calcium ions from the cross-link sites [18] [32] [67]. The process essentially "unmasks" the epitope by breaking the surrounding protein aggregates.
PIER: This enzymatic method relies on proteases such as proteinase K, trypsin, or pepsin to digest and degrade the proteins that constitute the cross-links [67] [21] [66]. By cleaving these peptide bonds, PIER clears a physical path for the antibody to access its target epitope.
Direct Comparison: HIER vs. PIER
The following table summarizes the core advantages and limitations of each method, providing a guide for initial method selection.
Table 1: Direct comparison of HIER and PIER methodologies.
| Feature | Heat-Induced Epitope Retrieval (HIER) | Proteolytic-Induced Epitope Retrieval (PIER) |
|---|---|---|
| Principle | Uses heat and buffer solutions to break cross-links [18] [32] | Uses enzymes (e.g., proteinase K, trypsin) to digest cross-links [67] [21] |
| Primary Use Case | Gentle, standard epitope retrieval; most common first-line approach [67] | Difficult epitope recovery; often used when HIER is ineffective [67] |
| Key Advantages | - Generally gentler on tissue morphology [18]- Wide applicability for many antigens [32]- Tunable via buffer pH and composition [18] [32] | - Can be highly effective for specific, hard-to-retrieve epitopes [8] [67]- Does not require specialized heating equipment |
| Key Limitations | - Requires precise control of time and temperature [18]- Can cause tissue section detachment, especially with high-pH buffers [18] [6]- Risk of damaging heat-labile epitopes [8] | - High risk of destroying tissue morphology and antigen if over-digested [6] [8]- Optimization of enzyme concentration and time is critical [21] |
| Typical Incubation | ~20 minutes at 95°C (varies by heat source) [6] [67] | ~10-20 minutes at 37°C [67] [21] |
| Critical Parameters | Buffer pH and composition, temperature, heating time, heating device [18] [32] | Enzyme type, concentration, incubation time, and temperature [21] |
Detailed Experimental Protocols
Standard HIER Protocol Using a Pressure Cooker
The pressure cooker is a widely used HIER heat source due to its even heat distribution and ability to reach high temperatures (110-120°C), allowing for shorter retrieval times [18] [6].
Materials Required:
- Domestic stainless steel pressure cooker
- Hot plate
- Slide rack and vessel
- Antigen retrieval buffer (e.g., Citrate pH 6.0, Tris-EDTA pH 9.0) [6]
Procedure:
- Deparaffinize and Rehydrate: Process slides through xylene and graded alcohols to water [6].
- Heat Buffer: Add a sufficient volume of antigen retrieval buffer to the pressure cooker, place it on a hot plate set to full power, and bring to a boil. Rest the lid on top but do not secure it [6].
- Transfer Slides: Once boiling, carefully transfer the slide rack from the tap water into the pressure cooker [6].
- Pressurize: Secure the pressure cooker lid as per the manufacturer's instructions. Once full pressure is reached, time for 3 minutes [6].
- Cool: Turn off the hotplate, place the cooker in a sink, activate the pressure release valve, and run cold water over it. Once de-pressurized, open the lid and run cold water into the cooker for 10 minutes to cool the slides [6].
- Continue Staining: Proceed with the subsequent steps of the IHC staining protocol, such as peroxidase blocking and antibody application [6].
Standard PIER Protocol Using Proteinase K
PIER is particularly useful for antigens in dense extracellular matrices, such as cartilage, where it may outperform HIER [8].
Materials Required:
- Proteinase K (e.g., 20 μg/mL working solution)
- Humidified incubation chamber
- TE Buffer, pH 8.0 [21]
Procedure:
- Deparaffinize and Rehydrate: Process slides to water as described in the HIER protocol.
- Apply Enzyme: Cover the tissue sections with the pre-warmed Proteinase K working solution [21].
- Digest: Incubate the slides for 10-20 minutes at 37°C in a humidified chamber to prevent evaporation [21].
- Stop Reaction: Remove the slides and transfer them to a rack in a container with tap water. Rinse in running water for several minutes to stop the enzymatic reaction [6] [21].
- Continue Staining: Proceed with the rest of the IHC protocol.
Workflow Visualization
The following diagram illustrates the decision pathway for selecting and optimizing an antigen retrieval method, integrating both HIER and PIER approaches.
The Scientist's Toolkit: Essential Reagents and Materials
Successful antigen retrieval requires careful selection of reagents and equipment. The following table lists key solutions and their functions.
Table 2: Key research reagent solutions for antigen retrieval.
| Item | Function and Application | Key Considerations |
|---|---|---|
| Citrate Buffer (pH 6.0) | A popular, mild retrieval buffer for a wide range of antigens [6] [32]. | Often the first buffer tested; provides a good balance of efficacy and morphology preservation [18]. |
| Tris-EDTA Buffer (pH 9.0) | High-pH buffer effective for many nuclear antigens and difficult-to-retrieve epitopes [6] [32]. | Can increase the risk of tissue section detachment from slides [18]. |
| EDTA Buffer (pH 8.0) | Effective for over-fixed specimens and challenging antigens; acts as a calcium chelator [18] [6]. | May cause distorted tissue morphology and convoluted nuclei [18]. |
| Proteinase K | A broad-spectrum serine protease used in PIER to digest protein cross-links [8] [21]. | Concentration and time must be tightly optimized to avoid destruction of tissue architecture and the antigen itself [8] [21]. |
| Trypsin | A protease commonly used in PIER protocols [67] [21]. | Typically used as a 0.05% solution at 37°C; pH must be adjusted to 7.8 for optimal activity [21]. |
| Pressure Cooker | A common and effective heat source for HIER, providing uniform heating and high temperatures [18] [6]. | Enables short incubation times (e.g., 2-5 min) but may cause tissue artifacts if not carefully controlled [18]. |
| Vegetable Steamer | A gentle and inexpensive heat source for HIER, maintaining temperatures of 95-100°C [18] [6]. | Requires longer heating times (e.g., 20 min) but is associated with good tissue morphology [18]. |
There is no universal "best" antigen retrieval method; the optimal choice is inherently antigen- and tissue-specific. As a guiding principle, HIER is the recommended first-line approach due to its broad efficacy and gentler impact on tissue morphology. However, for antigens embedded in dense matrices or those that are heat-labile, PIER may be a necessary and more effective alternative [8] [67]. A systematic, empirical comparison of both methods, as outlined in this note, is the cornerstone of a robust and optimized IHC protocol, ensuring reliable data for critical research and diagnostic decisions.
The dense extracellular matrix of articular cartilage presents a significant challenge for immunohistochemistry (IHC), particularly for detecting low-abundance glycoproteins with diagnostic potential for conditions like osteoarthritis. This application note details a systematic comparison of antigen retrieval methods, demonstrating that Proteolytic-Induced Epitope Retrieval (PIER) significantly outperforms Heat-Induced Epitope Retrieval (HIER) for the detection of Cartilage Intermediate Layer Protein 2 (CILP-2), a promising biomarker in osteoarthritis research [24] [8] [17]. The findings underscore that optimal antigen retrieval is not one-size-fits-all but must be adapted to the specific target protein and tissue type.
The study evaluated four different antigen retrieval protocols on osteoarthritic cartilage samples obtained from total knee replacement operations [24] [17]. A semi-quantitative assessment of CILP-2 staining extent revealed clear performance differences between the methods.
Table 1: Comparison of Antigen Retrieval Method Outcomes for CILP-2 IHC
| Antigen Retrieval Method | Protocol Summary | CILP-2 Staining Outcome | Key Observations |
|---|---|---|---|
| PIER | Proteinase K (30 µg/mL, 90 min, 37°C) + Hyaluronidase (0.4%, 3 h, 37°C) [8] [17] | Best / Most Abundant [24] [17] | Effective unmasking of epitopes in dense cartilage matrix [24]. |
| HIER | Reveal Decloaker buffer, 95°C for 10 min [8] [17] | Inferior to PIER [24] [17] | Potential destruction of antigenicity; suboptimal for CILP-2 [8]. |
| HIER/PIER Combination | Sequential HIER and PIER treatments [8] [17] | Reduced vs. PIER alone [24] [17] | Heat application reduced PIER's positive effect; caused frequent section detachment [24] [17]. |
| No Retrieval (Control) | Distilled water only [8] [17] | Least Effective [24] [17] | Confirms necessity of antigen retrieval for CILP-2 detection [24]. |
A critical finding was that combining HIER with PIER did not yield synergistic benefits. Instead, the application of heat reduced the positive effect of the enzymatic treatment and led to frequent detachment of tissue sections from the slides, compromising the integrity of the samples [24] [17].
Detailed Experimental Protocols
Sample Preparation
Cartilage biopsies were obtained from load-bearing sites of the tibial plateau of patients with end-stage knee osteoarthritis [8] [17]. Tissues were fixed in 10% buffered formalin for up to three weeks, decalcified, and embedded in paraffin. Sections were cut to a thickness of 4 µm and mounted on adhesion microscope slides [8] [17].
Antigen Retrieval Protocols
The following four protocols were compared on serial sections [8] [17]:
PIER Protocol:
HIER Protocol:
HIER/PIER Combination Protocol:
Control:
Immunohistochemistry Staining
Following antigen retrieval, all sections were processed uniformly [17]:
- Endogenous Peroxidase Blocking: 0.6% H₂O₂ for 15 minutes.
- Washing: 3 × 5 minutes in PBS (pH 7.4).
- Blocking: Dako REAL Antibody Diluent to block non-specific binding.
- Primary Antibody Incubation: Rabbit polyclonal anti-CILP-2 (Atlas Antibodies, cat no HPA041847) at a 1:100 dilution overnight at +4°C [17].
- Visualization: Dako REAL EnVision Detection System, Peroxidase/DAB+, Rabbit/Mouse, with DAB staining time adjusted for each antigen retrieval method [17] [68].
Diagram 1: Experimental workflow for comparing antigen retrieval methods in CILP-2 IHC.
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Materials and Reagents for CILP-2 IHC
| Item | Function / Application | Example Product / Specification |
|---|---|---|
| Anti-CILP-2 Antibody | Primary antibody for specific target detection | Rabbit polyclonal (e.g., Atlas Antibodies, HPA041847) [17] |
| Proteinase K | Proteolytic enzyme for PIER; digests cross-links | 30 µg/mL in 50 mM Tris/HCl, 5 mM CaCl₂, pH 6.0 [8] [17] |
| Hyaluronidase | Enzyme for PIER; degrades hyaluronic acid in cartilage matrix | 0.4% bovine hyaluronidase in HEPES buffer [8] [17] |
| HIER Buffer | Buffer for heat-induced epitope retrieval | Reveal Decloaker or similar (e.g., Citrate buffer pH 6.0, Tris-EDTA pH 9.0) [8] [69] |
| Detection System | Visualization of antibody binding | Dako REAL EnVision Detection System, Peroxidase/DAB+ [17] |
| Adhesion Microscope Slides | Prevents tissue detachment during rigorous PIER procedure | TOMO Adhesion Microscope Slides [8] [17] |
This case study demonstrates that PIER is the superior antigen retrieval method for the immunohistochemical detection of CILP-2 in osteoarthritic cartilage. The dense, glycosylated matrix of cartilage presents a formidable barrier that was more effectively overcome by enzymatic digestion with proteinase K and hyaluronidase than by heat-induced methods [24]. The failure of the HIER/PIER combination, which caused tissue detachment and reduced staining, highlights the delicate balance required in protocol optimization and cautions against assuming combinatorial methods are inherently better [24] [17].
The superior performance of PIER for CILP-2 may be attributed to the protein's biochemical properties. CILP-2 has fewer glycosylation sites than its isoform CILP-1, potentially making it more reliant on non-covalent bonds and thus more susceptible to heat-induced denaturation [8] [17]. Furthermore, the direct action of enzymes in breaking the protein cross-links formed during formalin fixation and digesting matrix components is particularly advantageous for a low-concentration protein embedded in a voluminous extracellular matrix [24] [70].
In conclusion, for researchers investigating minor matrix proteins in challenging tissues like articular cartilage, empirical testing of antigen retrieval protocols is indispensable. Relying solely on standard HIER protocols could lead to false negatives or an underestimation of protein presence. The data presented here provide a validated and detailed roadmap for optimizing IHC detection of CILP-2, thereby supporting more accurate research into its role in osteoarthritis pathogenesis and diagnosis.
Immunohistochemistry (IHC) serves as an indispensable technique in research and diagnostic pathology, enabling the visualization of protein localization within tissue architecture. A significant technical challenge, however, emerges from routine formalin fixation, which creates methylene cross-links between proteins, effectively masking antigenic epitopes and rendering them inaccessible to antibodies [6] [11]. Antigen retrieval (AR) was developed to reverse this masking and is now a cornerstone of modern IHC, critically influencing the success of detecting a wide array of targets [71]. The choice between the two primary AR methods—Heat-Induced Epitope Retrieval (HIER) and Proteolytic-Induced Epitope Retrieval (PIER)—is highly dependent on the subcellular localization of the target antigen and its biochemical environment. Within the context of optimizing AR methods, this application note provides targeted guidelines and detailed protocols to enhance the performance of antigen detection for nuclear, cytoplasmic, and membrane targets, ensuring reliable and reproducible results for researchers and drug development professionals.
HIER vs. PIER: Mechanisms and Comparative Analysis
Understanding the fundamental mechanisms of HIER and PIER is essential for selecting the appropriate method. Each technique employs a distinct strategy to break the cross-links formed during formalin fixation.
HIER uses high temperatures (typically 95-100°C) in conjunction with specific buffer solutions to hydrolyze methylene cross-links and restore the native conformation of epitopes [6]. The process may also involve calcium ion extraction and the unfolding of epitopes, making them accessible for antibody binding [6]. The pH of the retrieval buffer is a critical factor; commonly used buffers include sodium citrate (pH 6.0), Tris-EDTA (pH 9.0), and EDTA (pH 8.0) [6].
PIER, the earlier established method, relies on enzymatic digestion—using enzymes like Proteinase K, trypsin, or pepsin—to proteolytically degrade the protein crosslinks themselves, thereby freeing the epitopes [8] [71]. The effectiveness of PIER is highly dependent on enzyme concentration, incubation time, temperature, and the duration of initial fixation [8].
The decision to use HIER or PIER can significantly impact staining outcomes. The table below summarizes a comparative analysis based on recent studies.
Table 1: Comparative Analysis of HIER and PIER Performance
| Aspect | Heat-Induced Epitope Retrieval (HIER) | Proteolytic-Induced Epitope Retrieval (PIER) |
|---|---|---|
| Primary Mechanism | Hydrolytic cleavage of cross-links using heat and buffer [6] | Enzymatic digestion of cross-links [8] |
| Typical Conditions | 95-100°C for 10-20 min in citrate, Tris-EDTA, or EDTA buffer [6] | Proteinase K (e.g., 30 µg/mL, 37°C, 90 min) [8] |
| Tissue Morphology | Generally better preservation [15] | Higher risk of tissue damage, particularly with over-digestion [6] |
| Experimental Flexibility | Multiple devices available (pressure cooker, microwave, steamer) [6] | Protocol is generally simpler, requiring only an incubator [6] |
| Best for Specific Antigens | Membrane targets (e.g., HSV-2) and Nuclear targets [15] | Cytoplasmic/Matrix glycoproteins (e.g., CILP-2) [8] |
| Reported Performance | Superior for HSV-2 and eosinophil protein in vaginal tissue [15] | Most effective for CILP-2 in osteoarthritic cartilage; HIER reduced its efficacy [8] |
The following workflow diagram outlines the key decision points for selecting and optimizing an antigen retrieval method.
Antigen-Specific Retrieval Guidelines
The subcellular localization of a target antigen is a primary determinant for the choice of AR method. The distinct biochemical and structural milieus of different cellular compartments mean that no single AR method is universally optimal.
Nuclear Targets
Nuclear proteins, including transcription factors and cell cycle regulators, are often tightly bound to DNA within the chromatin structure. Formalin fixation adds an additional layer of cross-linking.
- Recommended Method: HIER is highly effective for nuclear antigens [15]. The combination of heat and appropriate buffer helps break the protein-DNA and protein-protein cross-links, efficiently exposing epitopes.
- Buffer Selection: A high-pH buffer, such as Tris-EDTA (pH 9.0), is often preferred as it aids in disrupting the strong ionic interactions within the nucleus.
- Protocol Consideration: Standard heating at 95-100°C for 10-20 minutes using a pressure cooker, microwave, or steamer is typically successful [6].
Cytoplasmic Targets
The cytoplasm contains a diverse array of proteins, including soluble enzymes, structural components, and proteins within dense extracellular matrices. The optimal AR method can be highly variable.
- General Recommendation: Both HIER and PIER can be effective, and empirical testing is required.
- Key Exception - Matrix Glycoproteins: For proteins residing in a voluminous and dense extracellular matrix, such as cartilage intermediate layer protein 2 (CILP-2), PIER has demonstrated superior performance [8]. In a direct comparison, HIER alone or combined with PIER did not improve CILP-2 staining and sometimes led to tissue detachment.
- PIER Protocol: A successful protocol for a cartilage glycoprotein involved treatment with 30 µg/mL Proteinase K in Tris/HCl buffer for 90 minutes at 37°C, followed by bovine hyaluronidase for 3 hours at 37°C [8]. This enzymatic combination effectively digests the dense matrix, allowing antibody penetration.
Membrane Targets
Membrane proteins, including receptors and viral coat proteins, are embedded in the phospholipid bilayer and can be challenging targets.
- Recommended Method: HIER is often the most suitable choice. Research on detecting the membrane-bound Herpes Simplex Virus-2 (HSV-2) in vaginal tissue found that HIER provided visibly increased antibody binding compared to other methods [15].
- Buffer Selection: Citrate buffer (pH 6.0) is a commonly used and effective buffer for many membrane targets.
- Protocol Insight: The cited study used heat retrieval at 80°C in citrate buffer for 20 minutes, which proved efficient for sample processing and subsequent quantitative analysis [15].
Table 2: Antigen Retrieval Recommendations Based on Target Localization
| Target Localization | Recommended Method | Optimal Buffer / Enzyme | Example Antigens & Performance Notes |
|---|---|---|---|
| Nuclear | HIER [15] | Tris-EDTA (pH 9.0) [6] | Transcription factors, Ki-67. High-pHI buffer effectively disrupts protein-DNA complexes. |
| Cytoplasmic / Matrix | PIER (preferred for matrix) [8] | Proteinase K (e.g., 30 µg/mL) [8] | CILP-2 (cartilage glycoprotein). PIER yielded most abundant staining; HIER reduced efficacy [8]. |
| Membrane | HIER [15] | Citrate Buffer (pH 6.0) [6] | HSV-2 viral coat protein. HIER at 80°C significantly increased antibody binding [15]. |
Detailed Experimental Protocols
Standardized HIER Protocol (Pressure Cooker Method)
This protocol is a robust and widely used approach for HIER, suitable for many nuclear and membrane targets [6].
Materials Required:
- Domestic stainless steel pressure cooker
- Hot plate
- Metal slide rack and vessel (~400-500 mL capacity)
- Antigen retrieval buffer (e.g., Citrate pH 6.0, Tris-EDTA pH 9.0)
Step-by-Step Procedure:
- Add a sufficient volume of antigen retrieval buffer to the pressure cooker to cover slides by several centimeters. Place the open cooker on a hot plate at full power.
- While the buffer is heating, deparaffinize and rehydrate the FFPE tissue sections using standard xylene and ethanol series.
- Once the buffer is boiling, carefully transfer the slides from the final water wash into the buffer in the pressure cooker using forceps.
- Secure the lid on the pressure cooker according to the manufacturer's instructions.
- Once full pressure is reached, start timing and maintain pressure for 3 minutes.
- After 3 minutes, turn off the hotplate and move the pressure cooker to a sink. Activate the pressure release valve and run cold water over the cooker to depressurize quickly.
- Open the lid and run cold tap water into the cooker for 10 minutes to cool the slides.
- Proceed with the subsequent steps of the IHC staining protocol (e.g., peroxidase blocking, antibody application).
Standardized PIER Protocol (Proteinase K Method)
This protocol is recommended for challenging cytoplasmic and extracellular matrix targets [8].
Materials Required:
- Proteinase K (e.g., Merck KGaA)
- Incubator or humidity chamber set to 37°C
- Tris/HCl buffer (50 mM, pH 6.0) with 5 mM CaCl₂
- Hyaluronidase (optional, for dense matrices) [8]
Step-by-Step Procedure:
- Deparaffinize and rehydrate the FFPE tissue sections.
- Prepare a working solution of 30 µg/mL Proteinase K in a Tris/HCl buffer (pH 6.0) containing 5 mM CaCl₂.
- Pipette the Proteinase K working solution onto the slides, ensuring complete coverage of the tissue.
- Place the slides in a humidity chamber and incubate for 90 minutes at 37°C.
- For dense matrices like cartilage, follow with a treatment of 0.4% bovine hyaluronidase in HEPES-buffered medium for 3 hours at 37°C [8].
- Rake the slides gently in Tris-buffered saline (TBS) or phosphate-buffered saline (PBS) to stop the enzymatic reaction.
- Proceed immediately with the remaining IHC steps.
The Scientist's Toolkit: Essential Reagents and Materials
Successful antigen retrieval relies on a set of core reagents and equipment. The following table details key solutions and their specific functions in the AR process.
Table 3: Essential Research Reagent Solutions for Antigen Retrieval
| Reagent / Material | Function and Application | Specific Examples & Notes |
|---|---|---|
| Citrate Buffer (pH 6.0) | A low-pH buffer for HIER; suitable for a wide range of antigens, particularly many membrane targets [6] [15]. | 10 mM Sodium citrate, 0.05% Tween 20. A versatile, first-choice buffer [6]. |
| Tris-EDTA Buffer (pH 9.0) | A high-pH buffer for HIER; effective for disrupting strong bonds, making it ideal for many nuclear targets [6]. | 10 mM Tris base, 1 mM EDTA, 0.05% Tween 20. Helps in breaking protein-nucleic acid cross-links [6]. |
| Proteinase K | A broad-spectrum serine protease used in PIER to digest protein cross-links, crucial for dense matrix and cytoplasmic targets [8]. | Working concentration of 30 µg/mL in Tris/HCl. Concentration and time must be optimized to avoid tissue damage [8]. |
| Hyaluronidase | An enzyme that digests hyaluronic acid, a major component of the extracellular matrix. Used in combination with proteases for complex tissues [8]. | 0.4% solution in HEPES buffer. Used after Proteinase K for enhanced retrieval in cartilage [8]. |
| Pressure Cooker / Decloaking Chamber | A device to achieve uniform and rapid heating of slides in buffer under pressure, ensuring consistent and robust HIER [6]. | Domestic pressure cookers are effective. Maintains temperature above 100°C, leading to shorter retrieval times (e.g., 3 min) [6]. |
The optimization of antigen retrieval is a critical, target-dependent process that directly determines the success of IHC experiments. As detailed in this application note, the fundamental guideline is to employ HIER for nuclear and membrane targets and strongly consider PIER for cytoplasmic proteins, particularly those embedded in dense extracellular matrices. The provided comparative data and detailed protocols serve as a foundational framework. However, given the unique nature of each antigen-antibody pair and tissue type, empirical optimization of buffer pH, enzyme concentration, and heating conditions remains an indispensable part of the workflow. Adhering to these antigen-specific guidelines will enable scientists to consistently generate high-quality, reliable IHC data, thereby accelerating research and drug development efforts.
Antigen retrieval is a critical step in immunohistochemistry (IHC) for unmasking epitopes concealed during formalin fixation and paraffin embedding [72]. This process enhances antibody binding efficiency, leading to stronger, more specific staining while minimizing background interference [73]. The two principal antigen retrieval methodologies are Heat-Induced Epitope Retrieval (HIER) and Proteolytic-Induced Epitope Retrieval (PIER) [28] [74]. A combined sequential approach (HIER + PIER) has been explored for challenging targets, but its applications are highly specific and not universally beneficial [8]. This application note evaluates the efficacy of the combined HIER + PIER approach, detailing its limited applications and providing optimized protocols for researchers and drug development professionals.
Mechanism of Action: HIER vs. PIER
Understanding the distinct mechanisms by which HIER and PIER unmask epitopes is fundamental to evaluating their combined use.
HIER (Heat-Induced Epitope Retrieval): This method utilizes heat, typically applied via a microwave, pressure cooker, or water bath, to reverse the methylene crosslinks formed between proteins during formalin fixation [72] [74]. The application of heat is believed to cause crosslinked proteins to unfold, restoring the secondary or tertiary structure of the epitope and making it accessible to antibody binding [28] [74].
PIER (Proteolytic-Induced Epitope Retrieval): As an earlier established method, PIER relies on enzymes such as trypsin, pepsin, or Proteinase K to cleave peptide bonds that may be physically masking the epitope [72] [41]. The mechanism involves the enzymatic degradation of protein crosslinks, thereby liberating the epitope for antibody interaction [8] [74].
The table below summarizes the core characteristics of each method.
Table 1: Fundamental Characteristics of HIER and PIER
| Feature | HIER | PIER |
|---|---|---|
| Principle | Heat-mediated reversal of protein crosslinks [74] | Enzymatic cleavage of masking peptides [74] |
| Common Agents | Heated buffer solutions (e.g., Sodium Citrate, Tris-EDTA) [72] | Proteolytic enzymes (e.g., Trypsin, Proteinase K, Pepsin) [72] [41] |
| Typical Conditions | 95-120°C for 1-20 minutes [28] [74] | 37°C for 10-20 minutes [41] |
| Primary Advantage | Higher success rate for most antigens; better tissue morphology preservation [73] [28] | Effective for certain masked epitopes less responsive to heat [8] [72] |
| Primary Disadvantage | Potential for tissue damage and epitope destruction if over-heated [8] | Risk of excessive tissue digestion and destruction of the target antigen [72] [28] |
Experimental Evaluation of a Combined HIER+PIER Approach
A direct comparison of antigen retrieval methods, including a combined HIER/PIER protocol, was conducted for detecting Cartilage Intermediate Layer Protein 2 (CILP-2) in human osteoarthritic cartilage, a tissue with a dense extracellular matrix that poses significant challenges for antibody penetration [8].
Key Experimental Findings
The study revealed that the combined HIER/PIER approach did not yield superior results. Specifically [8]:
- PIER alone produced the best staining results for CILP-2.
- The application of heat reduced the positive effect of PIER.
- Combining the methods led to practical issues, including the frequent detachment of tissue sections from the slides.
It was hypothesized that the lower glycosylation of CILP-2 compared to its isoform CILP-1 may contribute to its lower stability, making it more susceptible to heat-induced denaturation, thereby explaining the failure of the HIER-inclusive protocols [8].
Quantitative Staining Assessment
The semi-quantitative assessment of CILP-2 staining extent across the different protocols is summarized in the table below.
Table 2: Semi-Quantitative Staining Assessment for CILP-2 in OA Cartilage [8]
| Antigen Retrieval Method | Protocol Summary | Staining Outcome for CILP-2 |
|---|---|---|
| No Retrieval (Control) | Sections kept in distilled water without retrieval. | Lowest / No specific staining |
| HIER Only | Heat retrieval at 95°C for 10 min in Decloaker solution. | Weak staining |
| PIER Only | 30 µg/mL Proteinase K (90 min, 37°C) + 0.4% Hyaluronidase (3 h, 37°C). | Best staining results |
| HIER + PIER (Combined) | Sequential application of HIER and PIER protocols. | Reduced staining vs. PIER alone; section detachment |
Diagram 1: Antigen retrieval method decision pathway.
Detailed Protocols
Combined HIER + PIER Protocol (Based on CILP-2 Study)
The following protocol details the combined approach used in the CILP-2 study, which is recommended for contexts where PIER alone is insufficient and combination therapy is empirically determined to be beneficial [8].
Step 1: Deparaffinization and Rehydration
- Deparaffinize slides in xylene (2 changes, 5 min each).
- Rehydrate through graded ethanol series: 100% (2 changes), 95%, 70% (2 min each).
- Rise briefly in distilled water.
Step 2: Heat-Induced Epitope Retrieval (HIER)
- Place slides in a preheated Decloaker solution (e.g., Reveal Decloaker, Biocare Medical).
- Perform heat retrieval at 95°C for 10 minutes.
- Allow the slides to cool at room temperature for 20-30 minutes.
Step 3: Proteolytic-Induced Epitope Retrieval (PIER)
- Prepare Proteinase K working solution (20 µg/mL in TE Buffer, pH 8.0).
- Cover sections with the solution and incubate for 90 minutes at 37°C in a humidified chamber.
- Rinse slides with PBS.
- Treat sections with 0.4% bovine hyaluronidase in HEPES-buffered medium for 3 hours at 37°C [8].
Step 4: Immunohistochemical Staining
- Proceed with standard IHC staining: peroxide blocking, protein blocking, primary and secondary antibody incubation, detection, and counterstaining [8].
Standardized Individual Retrieval Protocols
Heat-Induced Epitope Retrieval (HIER) via Pressure Cooker
This is a robust and widely used HIER method [72].
- Step 1: Add antigen retrieval buffer (e.g., 10 mM Sodium Citrate, pH 6.0) to the pressure cooker. Heat until boiling.
- Step 2: After boiling, place deparaffinized and rehydrated slides into the cooker, close the lid tightly.
- Step 3: Once maximum pressure is reached, maintain it for 3 minutes.
- Step 4: Release steam, open the lid carefully, and cool the slides in cold water for 10 minutes.
Proteolytic-Induced Epitope Retrieval (PIER) using Trypsin
A common PIER protocol suitable for many antigens [72] [41].
- Step 1: Prepare Trypsin Working Solution (0.05%):
- 1 ml Trypsin stock solution (0.5%)
- 1 ml Calcium chloride stock solution (1%)
- 8 ml Distilled Water
- Adjust pH to 7.8 with 1N NaOH.
- Step 2: Cover the deparaffinized and rehydrated sections with the trypsin working solution.
- Step 3: Incubate for 10-20 minutes at 37°C in a humidified chamber.
- Step 4: Allow sections to cool at room temperature for 10 minutes before proceeding with IHC.
The Scientist's Toolkit: Research Reagent Solutions
Successful antigen retrieval requires specific reagents and tools. The following table lists essential materials and their functions.
Table 3: Essential Research Reagents for Antigen Retrieval
| Reagent / Tool | Function / Application | Example Protocols / Notes |
|---|---|---|
| Proteinase K | Proteolytic enzyme for PIER; cleaves peptide bonds to unmask epitopes. | Working solution: 20-30 µg/mL; incubation: 37°C for 10-90 min [8] [41]. |
| Trypsin | Proteolytic enzyme for a standard PIER protocol. | Working solution: 0.05%; incubation: 37°C for 10-20 min [72] [41]. |
| Hyaluronidase | Enzyme that digests hyaluronic acid; used in complex matrices like cartilage. | Used in combination with Proteinase K for cartilage samples (0.4%, 3h at 37°C) [8]. |
| Sodium Citrate Buffer (pH 6.0) | A common, mildly acidic retrieval buffer for HIER. | 10 mM Sodium Citrate, 0.05% Tween 20; compatible with most antibodies [72]. |
| Tris-EDTA Buffer (pH 9.0) | A common, basic retrieval buffer for HIER; often used for phospho-specific antibodies. | 10 mM Tris base, 1 mM EDTA, 0.05% Tween 20 [72]. |
| Pressure Cooker / Microwave | Apparatus for applying high, consistent heat during HIER. | Pressure cooker: 1-5 min at 120°C; Microwave: 20 min at 98°C [72] [28]. |
| Humidified Chamber | Essential for maintaining enzyme activity and solution consistency during PIER incubations. | Prevents evaporation of reagent solutions during 37°C incubation [72]. |
Optimization and Troubleshooting
Optimization Strategy for HIER
Optimizing HIER is empirical and requires testing a matrix of conditions. Key variables to test include [73] [28]:
- Buffer pH: Systematically test acidic (pH 6.0), neutral (pH 7.0-7.6), and basic (pH 8.0-9.0) buffers.
- Time and Temperature: Test incubation times (e.g., 1, 5, 15 minutes) at common temperatures (95-100°C for water baths, 120°C for pressure cookers).
- Validation: Always include a no-retrieval control and a known positive control tissue to validate results and identify artifacts [73].
Troubleshooting the Combined Approach
Problem: Tissue Section Detachment
- Cause: The sequential stress of heat and enzymatic digestion severely compromises tissue adhesion.
- Solution: Use high-quality adhesive slides. If detachment occurs, prioritize PIER alone or optimize HIER conditions to avoid the combination.
Problem: Weak or No Staining with Combined Method
- Cause: The antigen may be heat-labile (like CILP-2) and destroyed during the HIER step, or the epitope may be enzymatically cleaved by over-digestion in PIER.
- Solution: Systematically test HIER and PIER individually before combining. If the individual methods work but the combination fails, the antigen is likely not suitable for the combined approach.
Problem: Excessive Tissue Digestion
- Cause: Over-optimization of PIER incubation time or enzyme concentration.
- Solution: Titrate the enzyme concentration and reduce incubation time, monitoring tissue morphology closely.
Diagram 2: Troubleshooting weak staining in the combined approach.
The combined HIER + PIER antigen retrieval approach represents a specialized tool with narrow applicability. Empirical evidence demonstrates that it does not universally enhance staining and can be detrimental for specific targets, such as the cartilage glycoprotein CILP-2, leading to reduced immunoreactivity and technical failures like tissue detachment [8]. Therefore, researchers should prioritize systematic optimization of HIER and PIER as individual methods. The combined approach should be reserved for exceptionally challenging antigens where individual methods prove insufficient and is contingent upon rigorous empirical validation that confirms a clear benefit without compromising tissue integrity or antigenicity.
In immunohistochemistry (IHC), the reliability of your results hinges entirely on the specificity of your primary antibody. A poorly validated antibody can generate false-positive or false-negative data, leading to incorrect conclusions with significant scientific and clinical consequences. The International Working Group on Antibody Validation has established five conceptual pillars for antibody validation to address the reproducibility crisis in biomedical research, emphasizing that application-specific validation is critical [75]. While antigen retrieval methods like Heat-Induced Epitope Retrieval (HIER) and Proteolytic-Induced Epitope Retrieval (PIER) are crucial for optimizing epitope accessibility, these efforts are wasted if the antibody itself lacks specificity for its intended target.
This application note details rigorous validation strategies, provides protocols for integrating validation with antigen retrieval optimization, and presents a framework for ensuring your IHC data is both reliable and reproducible.
The Foundation: Linking Antigen Retrieval and Antibody Validation
The Dual Challenge of Epitope Masking and Antibody Specificity
Formalin fixation, the standard for tissue morphology preservation, creates methylene bridges between proteins, leading to epitope masking [2] [6]. Antigen retrieval (AR) is designed to reverse these cross-links, but the process itself introduces variability.
- AR Method Impact: The choice between HIER and PIER can dramatically affect antibody binding. HIER uses high temperature (95-120°C) and specific pH buffers to physically break cross-links, while PIER employs enzymes like trypsin or proteinase K to chemically digest masking proteins [76] [2].
- Validation Necessity: An antibody validated for IHC without AR may not function after HIER or PIER, as these processes can alter the epitope's conformation. Therefore, antibodies must be validated under the exact AR conditions used in the final protocol [77].
Key Validation Strategies for IHC
The table below outlines core antibody validation strategies endorsed by international guidelines, which can be used in combination for maximum confidence [75].
Table 1: Core Antibody Validation Strategies for IHC
| Validation Strategy | Core Principle | Key Advantage | Application in IHC |
|---|---|---|---|
| Orthogonal Validation | Cross-referencing IHC results with data from non-antibody methods (e.g., mass spectrometry, RNA-seq) [75] | Controls for antibody bias; provides independent confirmation | Compare IHC protein expression levels with transcriptomics data from sources like the Human Protein Atlas [75]. |
| Binary Validation | Testing antibody in systems with known positive and negative expression (e.g., knockout cells, siRNA) [75] | Directly confirms specificity by demonstrating absence of signal in negative controls | Use genetic knockouts or known positive/negative tissue controls to confirm specific staining is lost when the target is absent [75]. |
| Multiple Antibody Validation | Using multiple distinct antibodies against different epitopes on the same target | Confirms target identity by consistent staining pattern across different reagents | Different antibodies yielding the same staining pattern increases confidence in results. |
The following workflow integrates these validation strategies with antigen retrieval optimization for a robust IHC assay development process.
Advanced Validation: The Orthogonal Strategy
Definition and Implementation
Orthogonal validation involves corroborating antibody-based IHC results with data derived from methods that do not rely on antibodies [75]. This approach controls for antibody bias and provides independent verification of your experimental findings.
As explained by Katherine Crosby, Sr Director of Antibody Applications & Validation at Cell Signaling Technology, "Just as you need a different, calibrated weight to check if a scale is working correctly, you need antibody-independent data to cross-reference and verify the results of an antibody-driven experiment" [75].
Practical Workflow: Orthogonal Validation in Action
The diagram below illustrates a practical workflow for implementing orthogonal validation, using public 'omics data as a starting point.
Case Study: Validating a Nectin-2/CD112 Antibody
Researchers at Cell Signaling Technology validated their Nectin-2/CD112 antibody (clone D8D3F) using this exact workflow [75]:
- Orthogonal Data Mining: RNA expression data from the Human Protein Atlas was used to identify cell lines with high (RT4, MCF7) and low (HDLM-2, MOLT-4) expression of Nectin-2 [75].
- Binary Model Selection: These four cell lines were selected to create a system with known positive and negative expression [75].
- IHC Performance and Correlation: Western blot analysis (after optimized antigen retrieval) showed elevated expression in RT4 and MCF7 and minimal expression in HDLM-2 and MOLT-4, perfectly mirroring the orthogonal RNA data [75].
Table 2: Publicly Available Resources for Orthogonal Validation Data
| Resource | Data Type | Utility for IHC Validation |
|---|---|---|
| Human Protein Atlas [75] | Transcriptomics (RNA), Proteomics | Provides expected expression levels across tissues and cell lines; ideal for selecting positive/negative controls. |
| Cancer Cell Line Encyclopedia (CCLE) [75] | Genomic, Transcriptomic | Offers detailed genomic data for over 1,100 cancer cell lines to inform binary model selection. |
| COSMIC (Catalogue of Somatic Mutations In Cancer) [75] | Genomic, Mutation | Informs on mutation status of genes in cancers, which can predict protein expression levels. |
| DepMap Portal [75] | Multi-omics, Dependency | Integrates genomic and dependency data to help identify predictive models for validation. |
Experimental Protocols: Integrating Validation & Retrieval
Protocol: Optimizing Heat-Induced Epitope Retrieval (HIER)
HIER is the most widely used and successful AR method, utilizing high temperature to break formalin cross-links [78] [2]. This protocol is designed for systematic optimization.
Materials Required
- Antigen retrieval buffers: Sodium citrate (pH 6.0) and Tris-EDTA (pH 9.0) [6]
- Heating device (pressure cooker, water bath, or scientific microwave)
- Slide rack and Coplin jars or microwaveable vessel
Step-by-Step Method
- Deparaffinize and Rehydrate: Process paraffin-embedded tissue sections through xylene and graded alcohols to water [77].
- Select Retrieval Buffer: Begin testing with both a low-pH (e.g., Sodium citrate, pH 6.0) and a high-pH (e.g., Tris-EDTA, pH 9.0) buffer [78] [6].
- Apply Heat:
- Cool and Rinse: After heating, run cold tap water over the container for 10 minutes to cool slides and allow epitopes to re-nature [6].
- Proceed with Staining: Continue with the standard IHC protocol (blocking, primary antibody incubation, detection, etc.) [77].
Optimization Matrix A systematic matrix should be used to find the optimal HIER conditions for your specific antibody and tissue. Test different combinations of pH and incubation time while keeping temperature constant [78] [28].
Table 3: Experimental Matrix for Optimizing HIER Conditions
| Incubation Time | Antigen Retrieval Solution pH | ||
|---|---|---|---|
| Acidic (e.g., pH 6.0) | Neutral (e.g., pH 7.0) | Basic (e.g., pH 9.0) | |
| 1 minute | Slide #1 | Slide #2 | Slide #3 |
| 5 minutes | Slide #4 | Slide #5 | Slide #6 |
| 15 minutes | Slide #7 | Slide #8 | Slide #9 |
Compare all slides against a control with no HIER treatment to assess improvement [28].
Protocol: Incorporating Binary Controls for Validation
This protocol should be run in parallel with HIER optimization to confirm antibody specificity.
Materials Required
- Validated primary antibody
- Isotype control or no-primary antibody control
- Known positive and negative expression tissue sections or cell pellets (e.g., from CCLE or literature) [75]
- Knockout cell line (if available) is the gold standard
Step-by-Step Method
- Source Controls: Identify and procure tissues or cell lines with documented high and low/no expression of your target. Genetic knockouts are ideal negative controls [75].
- Parallel Processing: Embed control samples alongside test samples in the same paraffin block. Section and process all slides simultaneously to ensure identical treatment [79].
- Stain and Analyze: Perform IHC with optimized HIER conditions on the entire set.
- Interpret Results: The antibody is specific if staining is strong in positive controls and absent in negative controls. Any staining in the negative control indicates non-specific binding, necessitating further optimization or a different antibody [75].
Table 4: Essential Research Reagent Solutions for IHC Validation
| Reagent / Resource | Function / Purpose | Example Use Case |
|---|---|---|
| Validated Primary Antibodies | Specifically binds target antigen; the core reagent. | Choose antibodies validated for IHC, preferably with published validation data (e.g., KO validation, orthogonal) [75]. |
| HIER Buffers (Citrate, Tris-EDTA) | Unmask epitopes by breaking formalin cross-links under heat. | Sodium Citrate (pH 6.0) for many targets; Tris-EDTA (pH 9.0) for phosphorylated targets [6]. |
| PrEST Antigen (Matched Peptide) | Ultimate specificity control for blocking experiments. | Pre-incubate antibody with immunogen peptide; specific staining should be abolished [2]. |
| Cell Line Controls | Provide known positive/negative expression systems for binary validation. | Use cell lines with expression confirmed by RNA-seq (e.g., from Human Protein Atlas) [75]. |
| Automated IHC Platforms | Improve reproducibility and standardization of staining protocols. | Essential for clinical labs; reduces manual protocol variation [77] [79]. |
Rigorous antibody validation is not a standalone step but an integral part of developing a robust IHC assay. It must be performed in conjunction with the optimization of critical parameters like antigen retrieval. By adopting a multi-pronged validation strategy—incorporating orthogonal data, binary controls, and the systematic optimization of HIER—researchers can ensure their IHC data is specific, reproducible, and reliable.
As emphasized in the 2024 update to the College of American Pathologists guidelines, this rigorous approach is essential for both research and clinical applications to ensure accuracy and reduce variation in IHC practices [79]. In an era of increasing focus on scientific reproducibility, a thoroughly validated antibody is not just a best practice—it is the foundation of scientific integrity.
Conclusion
Optimizing antigen retrieval is not a one-size-fits-all endeavor but a strategic process vital for IHC reproducibility. The choice between HIER and PIER must be guided by the target antigen, tissue type, and fixation method, with HIER often serving as the preferred starting point for its broader applicability and gentler impact on morphology. However, as evidenced by specific cases like CILP-2 in cartilage, PIER remains indispensable for certain difficult-to-recover epitopes. A rigorous, systematic approach to optimization—incorporating methodical testing of buffers and conditions, comprehensive controls, and validated antibodies—is the cornerstone of success. Future directions point towards more standardized, automated protocols and a deeper investigation into the biochemical mechanisms of retrieval, which will further enhance the reliability of IHC in both fundamental research and clinical diagnostics, ultimately accelerating drug development and improving diagnostic precision.
References
- 1. Effects of Fixation and Tissue Processing on ... [https://www.leicabiosystems.com/us/knowledge-pathway/effects-of-fixation-and-tissue-processing-on-immunocytochemistry/]
- 2. Antigen Retrieval in IHC: Why It Matters and How to Get ... [https://www.atlasantibodies.com/knowledge-hub/blog/antigen-retrieval-in-ihc-why-it-matters-and-how-to-get-it-right/]
- 3. Antigen retrieval [https://en.wikipedia.org/wiki/Antigen_retrieval]
- 4. Antigen Masking During Fixation and Embedding, Dissected [https://pmc.ncbi.nlm.nih.gov/articles/PMC5256198/]
- 5. Effects of Prolonged Formalin Fixation on Diagnostic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2713075/]
- 6. IHC antigen retrieval protocol [https://www.abcam.com/en-us/technical-resources/protocols/ihc-antigen-retrieval]
- 7. Antigen retrieval and permeabilization for IHC [https://www.abcam.com/en-us/technical-resources/guides/ihc-guide/antigen-retrieval-and-permeabilization]
- 8. Comparison of Antigen Retrieval Methods for ... [https://www.mdpi.com/2409-9279/7/5/67]
- 9. IHC Epitope Retrieval (HIER vs. PIER) [https://www.novusbio.com/epitope-retrieval?srsltid=AfmBOoq6Xd1z37yH7BfJ-13blfke1iwSGepiKUDY7klReg48LZ0ALxQ8]
- 10. Optimizing HIER Antigen Retrieval for IHC [https://www.bosterbio.com/blog/post/protocol-for-optimizing-hier-antigen-retrieval-conditions?srsltid=AfmBOoqGnZ0GCKBLw5GvxDkFKewRYXrI746TOvDq3hcC5izImExa-hY7]
- 11. Immunohistochemistry (IHC): The Complete Guide [https://www.antibodies.com/applications/immunohistochemistry]
- 12. Molecular mechanisms of antigen retrieval - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC4676902/]
- 13. IHC Antigen Retrieval Protocol | Heat & Enzyme Methods [https://www.bosterbio.com/protocol-and-troubleshooting/ihc-optimization/antigen-retrieval?srsltid=AfmBOoolfx4R3RwcrdUyg1OOYjBzWnNUYgbuZ0Pl32229SITVI5HPk3u]
- 14. Evaluation and optimization of the immunohistochemistry ... [https://www.sciencedirect.com/science/article/abs/pii/S0022175923000066]
- 15. Walking a thin line between fixation and epitope binding [https://pmc.ncbi.nlm.nih.gov/articles/PMC11059462/]
- 16. Recent advances in antibody optimization based on deep ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12119181/]
- 17. Comparison of Antigen Retrieval Methods for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11417793/]
- 18. An Intro to Heat Induced Epitope Retrieval (HIER) in IHC [https://www.leicabiosystems.com/us/knowledge-pathway/technical-brief-of-heat-induced-epitope-retrieval/]
- 19. Antigen Retrieval-Should I do PIER or HIER? [https://www.sinobiological.com/category/antigen-retrieval-pier-or-hier]
- 20. IHC Epitope Retrieval (HIER vs. PIER) [https://www.novusbio.com/epitope-retrieval?srsltid=AfmBOoqrBavGbaQUGJjilgpQT8vU4UU8IqpcZc3dCxTm-fQzO7k-UlzR]
- 21. Antigen Retrieval Protocol (PIER) | Proteolytic-Induced ... [https://www.bio-techne.com/resources/protocols-troubleshooting/antigen-retrieval-protocol-pier]
- 22. Heat-induced Antigen Retrieval Protocol [https://www.rockland.com/resources/antigen-retrieval-methods-protocol/?srsltid=AfmBOoqVex8UyNAN-WgcUipRJLJA-fQ_le2kCiKyFRLrOPk2Aqv7Wrst]
- 23. Protocol for Heat-Induced Epitope Retrieval (HIER) [https://www.rndsystems.com/resources/protocols/protocol-heat-induced-epitope-retrieval-hier]
- 24. Comparison of Antigen Retrieval Methods for ... [https://pubmed.ncbi.nlm.nih.gov/39311368/]
- 25. Fresh frozen vs FFPE samples for next-generation ... [https://www.lexogen.com/blog/fresh-frozen-vs-ffpe-samples-for-next-generation-sequencing-studies/]
- 26. FFPE vs Frozen Tissue Samples [https://www.biochain.com/blog/ffpe-vs-frozen-tissue-samples/]
- 27. Frozen vs. Formalin-Fixed Paraffin-Embedded Tissue ... [https://www.stagebio.com/blog/frozen-vs-formalin-fixed-paraffin-embedded-tissue-sections-for-ihc]
- 28. Antigen Retrieval Methods [https://www.rndsystems.com/resources/protocols/antigen-retrieval-methods]
- 29. Optimizing HIER Antigen Retrieval for IHC [https://www.bosterbio.com/blog/post/protocol-for-optimizing-hier-antigen-retrieval-conditions?srsltid=AfmBOooRWktiIQXs7vGnjw7keYgAjtC1Iy_jDu1sPwYWqe3sJZgqEVe2]
- 30. Antigen Retrieval - an overview [https://www.sciencedirect.com/topics/medicine-and-dentistry/antigen-retrieval]
- 31. A Guide to Better IHC Staining: Steps, Troubleshooting, ... [https://www.celnovte.com/solution/a-guide-to-better-ihc-staining-steps-troubleshooting-and-optimization/]
- 32. Heat-Induced Antigen Retrieval for Immunohistochemical ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7604828/]
- 33. IHC Epitope Retrieval (HIER vs. PIER) [https://www.novusbio.com/epitope-retrieval?srsltid=AfmBOopsqAy_EOLof553G7_jqt_FCj9W8IlPCEb1q9PuWEfWIWSgRaj1]
- 34. Optimizing HIER Antigen Retrieval for IHC [https://www.bosterbio.com/blog/post/protocol-for-optimizing-hier-antigen-retrieval-conditions?srsltid=AfmBOood175UbRFzkwj4_2WMc5yZMl4nTBthp0OQzfvbwgN-UaT8XqqY]
- 35. Optimizing Your Antigen Retrieval Method - HIER vs PIER [https://www.bosterbio.com/blog/post/optimizing-your-antigen-retrieval-method-hier-vs-pier?srsltid=AfmBOopSSgrHoVycIGYP78jUYeC0wfpl4dvR6Wsqn0CmA-DHeWQR-uv8]
- 36. Optimizing Your Antigen Retrieval Method - HIER vs PIER [https://www.bosterbio.com/blog/post/optimizing-your-antigen-retrieval-method-hier-vs-pier?srsltid=AfmBOooPhIH291pRb4UtC1Fw1CqpsWSCTtDpVkPsu3LvMm8H_P8pCDU2]
- 37. IHC Antigen Retrieval Protocol | Heat & Enzyme Methods [https://www.bosterbio.com/protocol-and-troubleshooting/ihc-optimization/antigen-retrieval?srsltid=AfmBOorMIwf9mG3DiVlMRBIEkc58usV6A5iNqU3EnQXeXvXx34rn3cpF]
- 38. Optimizing HIER Antigen Retrieval for IHC [https://www.bosterbio.com/blog/post/protocol-for-optimizing-hier-antigen-retrieval-conditions?srsltid=AfmBOoq5eJqrKbMK25uVDTuvkK_ru-0Ykqg6WisOXvx0hvfxOidgN87p]
- 39. IHC Epitope Retrieval (HIER vs. PIER) [https://www.novusbio.com/epitope-retrieval?srsltid=AfmBOop4GMljNZrxaJ2hMk3Jc67NdowcO9oHkjuhH3Me4CmL5T_ns_E_]
- 40. The Breakdown: Enzymes and Epitope Retrieval [https://biocare.net/media/white-papers/the-breakdown-enzymes-and-epitope-retrieval/]
- 41. Antigen Retrieval Protocol (PIER) [https://www.novusbio.com/support-by-application/antigen-retrieval-protocol]
- 42. IHC Antigen Retrieval [https://www.ptglab.com/support/immunohistochemistry-protocol/ihc-antigen-retrieval/?srsltid=AfmBOoqqNri-WvsOKopRyjkP0-m2fT4CWjwjrIVsl-MUOsvRHzzogJJr]
- 43. IHC Epitope Retrieval (HIER vs. PIER) [https://www.novusbio.com/epitope-retrieval?srsltid=AfmBOorl6AI_bioN9r1pRHGx9jywaXWtPqM9YJYeUkgLukjWJUtcl7FX]
- 44. Antigen Retrieval Protocol (HIER) [https://www.novusbio.com/support/support-by-application/antigen-retrieval/protocol.html?srsltid=AfmBOop6pu9Sj_gJzW16UlY-iaU3MxUCjWRA8UWYTKCvzAVAOg5Zjb8B]
- 45. Optimizing Your Antigen Retrieval Method - HIER vs PIER [https://www.bosterbio.com/blog/post/optimizing-your-antigen-retrieval-method-hier-vs-pier?srsltid=AfmBOopPX4fxOdELzwTWpAAT-UYEOtVeaFEiCZpw_279IeSVlPiI5QIP]
- 46. IHC Epitope Retrieval (HIER vs. PIER) [https://www.novusbio.com/epitope-retrieval?srsltid=AfmBOop0M8SCbIH4jSmJR0to8dqt2hXlTG3IF2DmLB9An5fC9ePxvhZF]
- 47. Impact of antigen retrieval protocols on the ... [https://link.springer.com/article/10.1007/s00418-023-02187-4]
- 48. IHC Epitope Retrieval (HIER vs. PIER) [https://www.novusbio.com/epitope-retrieval?srsltid=AfmBOoo3yTqsroC0laNCez6soUHIAt2Xvh8suJGgAJxYPXdAUtMrSQuT]
- 49. Optimizing HIER Antigen Retrieval for IHC [https://www.bosterbio.com/blog/post/protocol-for-optimizing-hier-antigen-retrieval-conditions?srsltid=AfmBOoo8XMXB_MLFxg1wRzqvFW6duMoXitdDbTyRGTEXguestnADCvEY]
- 50. Optimizing Your Antigen Retrieval Method - HIER vs PIER [https://www.bosterbio.com/blog/post/optimizing-your-antigen-retrieval-method-hier-vs-pier?srsltid=AfmBOopB-jj9SicTJkDDO37RK86WtY13-Ur_bLdExG4gXIgVcFJjTTjN]
- 51. Chromogen-based Immunohistochemical Method for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3460359/]
- 52. Selecting an Optimal Positive IHC Control for Verifying ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6437345/]
- 53. IHC Epitope Retrieval (HIER vs. PIER) [https://www.novusbio.com/epitope-retrieval?srsltid=AfmBOoqKfLq6Xa1J1T5pVUjDTDRS5m7IxBsfo8sMMC-riTR9vf8fZe7D]
- 54. Antigen Retrieval in Immunohistochemistry [https://www.bosterbio.com/blog/post/antigen-retrieval-in-immunohistochemistry?srsltid=AfmBOorKxfoeLnTNitbvay18iQFLl5IsG4G7AV25noS3f6crH2NntgM0]
- 55. Optimizing Your Antigen Retrieval Method - HIER vs PIER [https://www.bosterbio.com/blog/post/optimizing-your-antigen-retrieval-method-hier-vs-pier?srsltid=AfmBOorsqeHvvKVJW3gcuUbxdIlx_RyC9cCkdWEaHNwOcOlcSt2pMkRr]
- 56. Optimizing Your Antigen Retrieval Method - HIER vs PIER [https://www.bosterbio.com/blog/post/optimizing-your-antigen-retrieval-method-hier-vs-pier?srsltid=AfmBOord-IMXc-s98Del1mpNxRVK5VQ7AJHFL1TZDO10CxAi7Xn-n1WP]
- 57. IHC Antigen Retrieval Protocol | Heat & Enzyme Methods [https://www.bosterbio.com/protocol-and-troubleshooting/ihc-optimization/antigen-retrieval?srsltid=AfmBOorjCiAAX6euLLAUI2LKL3HzHLHMloyvm0mB7od3hkoIA0a7nECt]
- 58. Optimizing HIER Antigen Retrieval for IHC [https://www.bosterbio.com/blog/post/protocol-for-optimizing-hier-antigen-retrieval-conditions?srsltid=AfmBOorVvwXV3kHWheRVbxFtaaDczJeBp1myThk4l6jc-tDQttHUEgjh]
- 59. IHC Epitope Retrieval (HIER vs. PIER) [https://www.novusbio.com/epitope-retrieval?srsltid=AfmBOoqB1K8gpc2IvsHYrEA4QVcFQREBFcLH99WEUraJ46xsvxG4oGhr]
- 60. Antigen Retrieval Protocol (HIER) [https://www.novusbio.com/support/support-by-application/antigen-retrieval/protocol.html?srsltid=AfmBOopLP3mU3-sh8otHfZEpyFSKk7iLSpZvYquzzHLg0331CHPKviv_]
- 61. Ten Approaches That Improve Immunostaining: A Review of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8836139/]
- 62. Review of immunohistochemistry techniques: Applications, ... [https://www.sciencedirect.com/science/article/abs/pii/S0740257024000467?utm_source=chatgpt.com]
- 63. Antigen Retrieval Protocol (HIER) [https://www.bio-techne.com/resources/protocols-troubleshooting/antigen-retrieval-hier]
- 64. A novel automated IHC staining system for quality control ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11864879/]
- 65. Optimizing HIER Antigen Retrieval for IHC [https://www.bosterbio.com/blog/post/protocol-for-optimizing-hier-antigen-retrieval-conditions?srsltid=AfmBOopIuApsG4WV7cpOSq6_NNybSp2Kh3p4FkxxFbQzLq2rGT5cNhnF]
- 66. IHC antigen retrieval: HIER . PIE vs [https://www.bio-techne.com/imaging/immunohistochemistry/epitope-retrieval]
- 67. IHC Antigen Retrieval | Proteintech Group [https://www.ptglab.com/support/immunohistochemistry-protocol/ihc-antigen-retrieval/]
- 68. Comparison of Antigen Retrieval Methods for ... [https://www.mdpi.com/2409-9279/7/5/67/review_report]
- 69. Optimizing HIER Antigen Retrieval for IHC [https://www.bosterbio.com/blog/post/protocol-for-optimizing-hier-antigen-retrieval-conditions?srsltid=AfmBOorZP0U5L_ydSwbp-DY8F5IeTHw5bQxfzdnwb926FL_Dtk82v8eL]
- 70. IHC Epitope Retrieval (HIER vs. PIER) [https://www.novusbio.com/epitope-retrieval?srsltid=AfmBOookBjRdQ94DzC-yWtdvtRcwfbdd5VELwIjYj8-MgOY7EzETjAJR]
- 71. Antigen Retrieval Immunohistochemistry [https://pmc.ncbi.nlm.nih.gov/articles/PMC3201121/]
- 72. IHC Antigen Retrieval Protocol [https://www.creative-diagnostics.com/ihc-antigen-retrieval-protocol.htm]
- 73. Optimizing HIER Antigen Retrieval for IHC [https://www.bosterbio.com/blog/post/protocol-for-optimizing-hier-antigen-retrieval-conditions?srsltid=AfmBOorXb1Q4D9CSgiUnjIkpw_y7Ri85d-2RMesLvJsI8w_I-RRSjvq-]
- 74. IHC Epitope Retrieval (HIER vs. PIER) [https://www.novusbio.com/epitope-retrieval?srsltid=AfmBOoqk0UL4l3NYIzTRNF1GSNdJGuD9cWQWh2I-ZJlv5p8Ync8_7k_V]
- 75. Antibody Validation Essentials: Orthogonal Strategy - CST Blog [https://blog.cellsignal.com/hallmarks-of-validation-orthogonal-strategy]
- 76. IHC Epitope Retrieval (HIER vs. PIER) [https://www.novusbio.com/epitope-retrieval?srsltid=AfmBOooHJ5nebx8YE1yeMLlXMhbkUpXD6KvRDqitK_7Hkw-u66OZ8vDN]
- 77. Immunohistochemistry for Pathologists: Protocols, Pitfalls, ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5122731/]
- 78. Optimizing HIER Antigen Retrieval for IHC [https://www.bosterbio.com/blog/post/protocol-for-optimizing-hier-antigen-retrieval-conditions?srsltid=AfmBOopz0OXqYmxcNCoLogmuLQhRBqaKcc6fAYomOyZprWITkf-1YmRQ]
- 79. Principles of Analytic Validation of Immunohistochemical ... [https://www.cap.org/protocols-and-guidelines/cap-guidelines/current-cap-guidelines/principles-of-analytic-validation-of-immunohistochemical-assays]